Integrated View of Baseline Protein Expression in Human Tissues
- PMID: 36577097
- PMCID: PMC9990129
- DOI: 10.1021/acs.jproteome.2c00406
Integrated View of Baseline Protein Expression in Human Tissues
Abstract
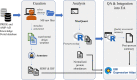
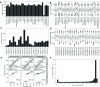
The availability of proteomics datasets in the public domain, and in the PRIDE database, in particular, has increased dramatically in recent years. This unprecedented large-scale availability of data provides an opportunity for combined analyses of datasets to get organism-wide protein abundance data in a consistent manner. We have reanalyzed 24 public proteomics datasets from healthy human individuals to assess baseline protein abundance in 31 organs. We defined tissue as a distinct functional or structural region within an organ. Overall, the aggregated dataset contains 67 healthy tissues, corresponding to 3,119 mass spectrometry runs covering 498 samples from 489 individuals. We compared protein abundances between different organs and studied the distribution of proteins across these organs. We also compared the results with data generated in analogous studies. Additionally, we performed gene ontology and pathway-enrichment analyses to identify organ-specific enriched biological processes and pathways. As a key point, we have integrated the protein abundance results into the resource Expression Atlas, where they can be accessed and visualized either individually or together with gene expression data coming from transcriptomics datasets. We believe this is a good mechanism to make proteomics data more accessible for life scientists.
Keywords: human proteome; mass spectrometry; public data re-use; quantitative proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Integrated View of Baseline Protein Expression in Human Tissues Using Public Data Independent Acquisition Data Sets.J Proteome Res. 2025 Feb 7;24(2):685-695. doi: 10.1021/acs.jproteome.4c00788. Epub 2025 Jan 7. J Proteome Res. 2025. PMID: 39764611 Free PMC article.
-
Integrated Proteomics Analysis of Baseline Protein Expression in Pig Tissues.J Proteome Res. 2024 Jun 7;23(6):1948-1959. doi: 10.1021/acs.jproteome.3c00741. Epub 2024 May 8. J Proteome Res. 2024. PMID: 38717300 Free PMC article.
-
Implementing the reuse of public DIA proteomics datasets: from the PRIDE database to Expression Atlas.Sci Data. 2022 Jun 14;9(1):335. doi: 10.1038/s41597-022-01380-9. Sci Data. 2022. PMID: 35701420 Free PMC article.
-
Mass Spectrometry-Based Chemical and Enzymatic Methods for Global Analysis of Protein Glycosylation.Acc Chem Res. 2018 Aug 21;51(8):1796-1806. doi: 10.1021/acs.accounts.8b00200. Epub 2018 Jul 16. Acc Chem Res. 2018. PMID: 30011186 Free PMC article. Review.
-
Approaches for systematic proteome exploration.Biomol Eng. 2007 Jun;24(2):155-68. doi: 10.1016/j.bioeng.2007.01.001. Epub 2007 Feb 6. Biomol Eng. 2007. PMID: 17376740 Review.
Cited by
-
Integrated View of Baseline Protein Expression in Human Tissues Using Public Data Independent Acquisition Data Sets.J Proteome Res. 2025 Feb 7;24(2):685-695. doi: 10.1021/acs.jproteome.4c00788. Epub 2025 Jan 7. J Proteome Res. 2025. PMID: 39764611 Free PMC article.
-
Expression Atlas update: insights from sequencing data at both bulk and single cell level.Nucleic Acids Res. 2024 Jan 5;52(D1):D107-D114. doi: 10.1093/nar/gkad1021. Nucleic Acids Res. 2024. PMID: 37992296 Free PMC article.
-
Integrated Proteomics Analysis of Baseline Protein Expression in Pig Tissues.J Proteome Res. 2024 Jun 7;23(6):1948-1959. doi: 10.1021/acs.jproteome.3c00741. Epub 2024 May 8. J Proteome Res. 2024. PMID: 38717300 Free PMC article.
-
Expression Atlas update: gene and protein expression in multiple species.Nucleic Acids Res. 2022 Jan 7;50(D1):D129-D140. doi: 10.1093/nar/gkab1030. Nucleic Acids Res. 2022. PMID: 34850121 Free PMC article.
-
Surprising features of nuclear receptor interaction networks revealed by live-cell single-molecule imaging.Elife. 2025 Jan 10;12:RP92979. doi: 10.7554/eLife.92979. Elife. 2025. PMID: 39792435 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

