Decreasing mutant ATXN1 nuclear localization improves a spectrum of SCA1-like phenotypes and brain region transcriptomic profiles
- PMID: 36577403
- PMCID: PMC9957934
- DOI: 10.1016/j.neuron.2022.11.017
Decreasing mutant ATXN1 nuclear localization improves a spectrum of SCA1-like phenotypes and brain region transcriptomic profiles
Abstract
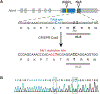
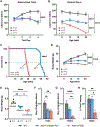
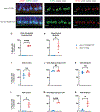
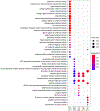
Spinocerebellar ataxia type 1 (SCA1) is a dominant trinucleotide repeat neurodegenerative disease characterized by motor dysfunction, cognitive impairment, and premature death. Degeneration of cerebellar Purkinje cells is a frequent and prominent pathological feature of SCA1. We previously showed that transport of ATXN1 to Purkinje cell nuclei is required for pathology, where mutant ATXN1 alters transcription. To examine the role of ATXN1 nuclear localization broadly in SCA1-like disease pathogenesis, CRISPR-Cas9 was used to develop a mouse with an amino acid alteration (K772T) in the nuclear localization sequence of the expanded ATXN1 protein. Characterization of these mice indicates that proper nuclear localization of mutant ATXN1 contributes to many disease-like phenotypes including motor dysfunction, cognitive deficits, and premature lethality. RNA sequencing analysis of genes with expression corrected to WT levels in Atxn1175QK772T/2Q mice indicates that transcriptomic aspects of SCA1 pathogenesis differ between the cerebellum, brainstem, cerebral cortex, hippocampus, and striatum.
Keywords: SCA1; neurodegeneration; nuclear localization; spinocerebellar ataxia type 1.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Comment in
-
Diverse regional mechanisms drive spinocerebellar ataxia type 1 phenotypes.Neuron. 2023 Feb 15;111(4):447-449. doi: 10.1016/j.neuron.2023.01.021. Neuron. 2023. PMID: 36796325
Comment on
-
Disruption of the ATXN1-CIC complex reveals the role of additional nuclear ATXN1 interactors in spinocerebellar ataxia type 1.Neuron. 2023 Feb 15;111(4):481-492.e8. doi: 10.1016/j.neuron.2022.11.016. Epub 2022 Dec 27. Neuron. 2023. PMID: 36577402 Free PMC article.
References
-
- Nucifora FC, Ellerby LM, Wellington CL, Wood JD, Herring WJ, Sawa A, Hayden MR, Dawson VL, Dawson TM, and Ross CA (2003). Nuclear localization of a non-caspase truncation product of atrophin-1, with an expanded polyglutamine repeat, increases cellular toxicity. J. Biol. Chem 278, 13047–13055. 10.1074/jbc.M211224200. - DOI - PubMed
-
- Bichelmeier U, Schmidt T, Hübener J, Boy J, Rüttiger L, Häbig K, Poths S, Bonin M, Knipper M, Schmidt WJ, et al. (2007). Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: In vivo evidence. J. Neurosci 27, 7418–7428. 10.1523/JNEUROSCI.4540-06.2007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

