Protein acylation: mechanisms, biological functions and therapeutic targets
- PMID: 36577755
- PMCID: PMC9797573
- DOI: 10.1038/s41392-022-01245-y
Protein acylation: mechanisms, biological functions and therapeutic targets
Abstract
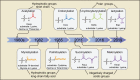
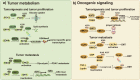
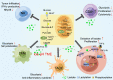
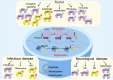
Metabolic reprogramming is involved in the pathogenesis of not only cancers but also neurodegenerative diseases, cardiovascular diseases, and infectious diseases. With the progress of metabonomics and proteomics, metabolites have been found to affect protein acylations through providing acyl groups or changing the activities of acyltransferases or deacylases. Reciprocally, protein acylation is involved in key cellular processes relevant to physiology and diseases, such as protein stability, protein subcellular localization, enzyme activity, transcriptional activity, protein-protein interactions and protein-DNA interactions. Herein, we summarize the functional diversity and mechanisms of eight kinds of nonhistone protein acylations in the physiological processes and progression of several diseases. We also highlight the recent progress in the development of inhibitors for acyltransferase, deacylase, and acylation reader proteins for their potential applications in drug discovery.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

