Human-in-the-loop assisted de novo molecular design
- PMID: 36578043
- PMCID: PMC9795720
- DOI: 10.1186/s13321-022-00667-8
Human-in-the-loop assisted de novo molecular design
Abstract
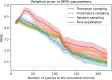
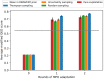
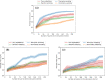
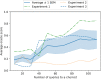
A de novo molecular design workflow can be used together with technologies such as reinforcement learning to navigate the chemical space. A bottleneck in the workflow that remains to be solved is how to integrate human feedback in the exploration of the chemical space to optimize molecules. A human drug designer still needs to design the goal, expressed as a scoring function for the molecules that captures the designer's implicit knowledge about the optimization task. Little support for this task exists and, consequently, a chemist usually resorts to iteratively building the objective function of multi-parameter optimization (MPO) in de novo design. We propose a principled approach to use human-in-the-loop machine learning to help the chemist to adapt the MPO scoring function to better match their goal. An advantage is that the method can learn the scoring function directly from the user's feedback while they browse the output of the molecule generator, instead of the current manual tuning of the scoring function with trial and error. The proposed method uses a probabilistic model that captures the user's idea and uncertainty about the scoring function, and it uses active learning to interact with the user. We present two case studies for this: In the first use-case, the parameters of an MPO are learned, and in the second use-case a non-parametric component of the scoring function to capture human domain knowledge is developed. The results show the effectiveness of the methods in two simulated example cases with an oracle, achieving significant improvement in less than 200 feedback queries, for the goals of a high QED score and identifying potent molecules for the DRD2 receptor, respectively. We further demonstrate the performance gains with a medicinal chemist interacting with the system.
Keywords: AI-assisted design; De novo molecular design; Expert knowledge elicitation; Goal-oriented molecule generation; Human-in-the-loop; Interactive algorithms; Reward elicitation.
© 2022. The Author(s).
Conflict of interest statement
This work was financially supported by AstraZeneca. The authors declare no other competing interests.
Figures


References
-
- Mervin L, Genheden S, Engkvist O. AI for drug design: From explicit rules to deep learning. Artif Intell Life Sci. 2022;2:100041. doi: 10.1016/J.AILSCI.2022.100041. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

