Alterations of oral microbiota and impact on the gut microbiome in type 1 diabetes mellitus revealed by integrated multi-omic analyses
- PMID: 36578059
- PMCID: PMC9795701
- DOI: 10.1186/s40168-022-01435-4
Alterations of oral microbiota and impact on the gut microbiome in type 1 diabetes mellitus revealed by integrated multi-omic analyses
Abstract
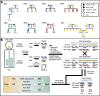
Background: Alterations to the gut microbiome have been linked to multiple chronic diseases. However, the drivers of such changes remain largely unknown. The oral cavity acts as a major route of exposure to exogenous factors including pathogens, and processes therein may affect the communities in the subsequent compartments of the gastrointestinal tract. Here, we perform strain-resolved, integrated meta-genomic, transcriptomic, and proteomic analyses of paired saliva and stool samples collected from 35 individuals from eight families with multiple cases of type 1 diabetes mellitus (T1DM).
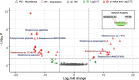
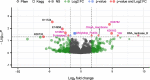
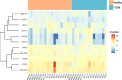
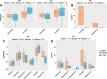
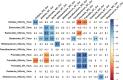
Results: We identified distinct oral microbiota mostly reflecting competition between streptococcal species. More specifically, we found a decreased abundance of the commensal Streptococcus salivarius in the oral cavity of T1DM individuals, which is linked to its apparent competition with the pathobiont Streptococcus mutans. The decrease in S. salivarius in the oral cavity was also associated with its decrease in the gut as well as higher abundances in facultative anaerobes including Enterobacteria. In addition, we found evidence of gut inflammation in T1DM as reflected in the expression profiles of the Enterobacteria as well as in the human gut proteome. Finally, we were able to follow transmitted strain-variants from the oral cavity to the gut at the individual omic levels, highlighting not only the transfer, but also the activity of the transmitted taxa along the gastrointestinal tract.
Conclusions: Alterations of the oral microbiome in the context of T1DM impact the microbial communities in the lower gut, in particular through the reduction of "mouth-to-gut" transfer of Streptococcus salivarius. Our results indicate that the observed oral-cavity-driven gut microbiome changes may contribute towards the inflammatory processes involved in T1DM. Through the integration of multi-omic analyses, we resolve strain-variant "mouth-to-gut" transfer in a disease context. Video Abstract.
© 2022. The Author(s).
Conflict of interest statement
All authors declare that they have no competing interests.
Figures
Similar articles
-
Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes.Nat Microbiol. 2016 Oct 10;2:16180. doi: 10.1038/nmicrobiol.2016.180. Nat Microbiol. 2016. PMID: 27723761
-
Unravelling the Role of Gut and Oral Microbiota in the Pediatric Population with Type 1 Diabetes Mellitus.Int J Mol Sci. 2024 Oct 2;25(19):10611. doi: 10.3390/ijms251910611. Int J Mol Sci. 2024. PMID: 39408940 Free PMC article. Review.
-
Bifidobacterium reduction is associated with high blood pressure in children with type 1 diabetes mellitus.Biomed Pharmacother. 2021 Aug;140:111736. doi: 10.1016/j.biopha.2021.111736. Epub 2021 May 23. Biomed Pharmacother. 2021. PMID: 34034069
-
Altered infective competence of the human gut microbiome in COVID-19.Microbiome. 2023 Mar 9;11(1):46. doi: 10.1186/s40168-023-01472-7. Microbiome. 2023. PMID: 36894986 Free PMC article.
-
The Oral and Gut Bacterial Microbiomes: Similarities, Differences, and Connections.Biol Res Nurs. 2021 Jan;23(1):7-20. doi: 10.1177/1099800420941606. Epub 2020 Jul 21. Biol Res Nurs. 2021. PMID: 32691605 Free PMC article. Review.
Cited by
-
The Complicated Relationship of Short-Chain Fatty Acids and Oral Microbiome: A Narrative Review.Biomedicines. 2023 Oct 11;11(10):2749. doi: 10.3390/biomedicines11102749. Biomedicines. 2023. PMID: 37893122 Free PMC article. Review.
-
Unveiling the connection between gut microbiome and metabolic health in individuals with chronic spinal cord injury.Physiol Genomics. 2024 Apr 1;56(4):317-326. doi: 10.1152/physiolgenomics.00107.2023. Epub 2024 Feb 12. Physiol Genomics. 2024. PMID: 38344780 Free PMC article.
-
Oral Microbiome Traits of Type 1 Diabetes and Phenylketonuria Patients in Latvia.Microorganisms. 2023 May 31;11(6):1471. doi: 10.3390/microorganisms11061471. Microorganisms. 2023. PMID: 37374973 Free PMC article.
-
The forgotten link: how the oral microbiome shapes childhood growth and development.Front Oral Health. 2025 Feb 7;6:1547099. doi: 10.3389/froh.2025.1547099. eCollection 2025. Front Oral Health. 2025. PMID: 39989601 Free PMC article. Review.
-
Omics for deciphering oral microecology.Int J Oral Sci. 2024 Jan 9;16(1):2. doi: 10.1038/s41368-023-00264-x. Int J Oral Sci. 2024. PMID: 38195684 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

