SARS-CoV-2 replication in airway epithelia requires motile cilia and microvillar reprogramming
- PMID: 36580912
- PMCID: PMC9715480
- DOI: 10.1016/j.cell.2022.11.030
SARS-CoV-2 replication in airway epithelia requires motile cilia and microvillar reprogramming
Abstract
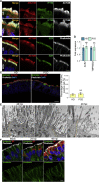
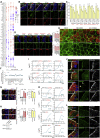
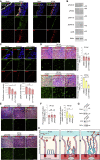
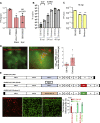
How SARS-CoV-2 penetrates the airway barrier of mucus and periciliary mucins to infect nasal epithelium remains unclear. Using primary nasal epithelial organoid cultures, we found that the virus attaches to motile cilia via the ACE2 receptor. SARS-CoV-2 traverses the mucus layer, using motile cilia as tracks to access the cell body. Depleting cilia blocks infection for SARS-CoV-2 and other respiratory viruses. SARS-CoV-2 progeny attach to airway microvilli 24 h post-infection and trigger formation of apically extended and highly branched microvilli that organize viral egress from the microvilli back into the mucus layer, supporting a model of virus dispersion throughout airway tissue via mucociliary transport. Phosphoproteomics and kinase inhibition reveal that microvillar remodeling is regulated by p21-activated kinases (PAK). Importantly, Omicron variants bind with higher affinity to motile cilia and show accelerated viral entry. Our work suggests that motile cilia, microvilli, and mucociliary-dependent mucus flow are critical for efficient virus replication in nasal epithelia.
Keywords: COVID-19; SARS-CoV-2; airway epithelial cells; cilia; microvilli; organoids; protein kinases; proteomics; respiratory synctital virus; respiratory virus.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures







Comment in
-
Across the mucus.Nat Rev Microbiol. 2023 Feb;21(2):65. doi: 10.1038/s41579-022-00842-6. Nat Rev Microbiol. 2023. PMID: 36513767 Free PMC article.
-
Motile cilia and microvillar: accomplices of SARS-CoV-2 in penetrating mucus barrier and infecting airway epithelium.Signal Transduct Target Ther. 2023 Mar 14;8(1):117. doi: 10.1038/s41392-023-01387-7. Signal Transduct Target Ther. 2023. PMID: 36918546 Free PMC article. No abstract available.
References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
-
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
- N01 AI040085/AI/NIAID NIH HHS/United States
- DP2 AI171139/AI/NIAID NIH HHS/United States
- R01 AI036178/AI/NIAID NIH HHS/United States
- R01 HD085901/HD/NICHD NIH HHS/United States
- T32 GM007276/GM/NIGMS NIH HHS/United States
- F32 GM142180/GM/NIGMS NIH HHS/United States
- U54 CA209971/CA/NCI NIH HHS/United States
- R01 GM121565/GM/NIGMS NIH HHS/United States
- R03 TR004209/TR/NCATS NIH HHS/United States
- R01 AI149672/AI/NIAID NIH HHS/United States
- P01 AI091575/AI/NIAID NIH HHS/United States
- S10 RR026780/RR/NCRR NIH HHS/United States
- R01 GM114276/GM/NIGMS NIH HHS/United States
- R01 DK127665/DK/NIDDK NIH HHS/United States
- P30 DK116074/DK/NIDDK NIH HHS/United States
- R01 CA250534/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

