Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants
- PMID: 36580913
- PMCID: PMC9747694
- DOI: 10.1016/j.cell.2022.12.018
Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants
Abstract
The BQ and XBB subvariants of SARS-CoV-2 Omicron are now rapidly expanding, possibly due to altered antibody evasion properties deriving from their additional spike mutations. Here, we report that neutralization of BQ.1, BQ.1.1, XBB, and XBB.1 by sera from vaccinees and infected persons was markedly impaired, including sera from individuals boosted with a WA1/BA.5 bivalent mRNA vaccine. Titers against BQ and XBB subvariants were lower by 13- to 81-fold and 66- to 155-fold, respectively, far beyond what had been observed to date. Monoclonal antibodies capable of neutralizing the original Omicron variant were largely inactive against these new subvariants, and the responsible individual spike mutations were identified. These subvariants were found to have similar ACE2-binding affinities as their predecessors. Together, our findings indicate that BQ and XBB subvariants present serious threats to current COVID-19 vaccines, render inactive all authorized antibodies, and may have gained dominance in the population because of their advantage in evading antibodies.
Keywords: BQ.1; BQ.1.1; COVID-19; SARS-CoV-2; XBB; XBB.1; antibody evasion; mRNA vaccine; neutralizing monoclonal antibody; receptor binding affinity.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.I, J.Y., Lihong Liu, and D.D.H. are inventors on patent applications (WO2021236998) or provisional patent applications (63/271,627) filed by Columbia University for a number of SARS-CoV-2 neutralizing antibodies described in this manuscript. Both sets of applications are under review. D.D.H. is a co-founder of TaiMed Biologics and RenBio, consultant to WuXi Biologics and Brii Biosciences, and board director for Vicarious Surgical. Aubree Gordon serves on a scientific advisory board for Janssen Pharmaceuticals. Other authors declare no competing interests.
Figures


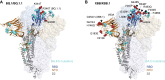




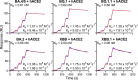
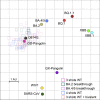
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

