Computational Methods for Single-cell Multi-omics Integration and Alignment
- PMID: 36581065
- PMCID: PMC10025765
- DOI: 10.1016/j.gpb.2022.11.013
Computational Methods for Single-cell Multi-omics Integration and Alignment
Abstract
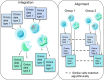
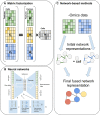
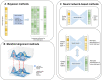
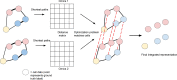
Recently developed technologies to generate single-cell genomic data have made a revolutionary impact in the field of biology. Multi-omics assays offer even greater opportunities to understand cellular states and biological processes. The problem of integrating different omics data with very different dimensionality and statistical properties remains, however, quite challenging. A growing body of computational tools is being developed for this task, leveraging ideas ranging from machine translation to the theory of networks, and represents another frontier on the interface of biology and data science. Our goal in this review is to provide a comprehensive, up-to-date survey of computational techniques for the integration of single-cell multi-omics data, while making the concepts behind each algorithm approachable to a non-expert audience.
Keywords: Integration; Machine learning; Multi-omics; Single-cell; Unsupervised learning.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
The authors have declared no competing interests.
Figures
References
-
- Peterson V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C., et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

