The sirtuin family in health and disease
- PMID: 36581622
- PMCID: PMC9797940
- DOI: 10.1038/s41392-022-01257-8
The sirtuin family in health and disease
Abstract
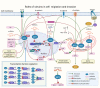
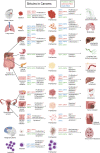
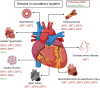
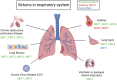
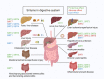
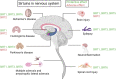
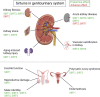
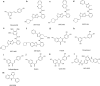
Sirtuins (SIRTs) are nicotine adenine dinucleotide(+)-dependent histone deacetylases regulating critical signaling pathways in prokaryotes and eukaryotes, and are involved in numerous biological processes. Currently, seven mammalian homologs of yeast Sir2 named SIRT1 to SIRT7 have been identified. Increasing evidence has suggested the vital roles of seven members of the SIRT family in health and disease conditions. Notably, this protein family plays a variety of important roles in cellular biology such as inflammation, metabolism, oxidative stress, and apoptosis, etc., thus, it is considered a potential therapeutic target for different kinds of pathologies including cancer, cardiovascular disease, respiratory disease, and other conditions. Moreover, identification of SIRT modulators and exploring the functions of these different modulators have prompted increased efforts to discover new small molecules, which can modify SIRT activity. Furthermore, several randomized controlled trials have indicated that different interventions might affect the expression of SIRT protein in human samples, and supplementation of SIRT modulators might have diverse impact on physiological function in different participants. In this review, we introduce the history and structure of the SIRT protein family, discuss the molecular mechanisms and biological functions of seven members of the SIRT protein family, elaborate on the regulatory roles of SIRTs in human disease, summarize SIRT inhibitors and activators, and review related clinical studies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Cellular and molecular biology of sirtuins in cardiovascular disease.Biomed Pharmacother. 2023 Aug;164:114931. doi: 10.1016/j.biopha.2023.114931. Epub 2023 May 30. Biomed Pharmacother. 2023. PMID: 37263163 Review.
-
A review on SIRT3 and its natural small molecule activators as a potential Preventive and therapeutic target.Eur J Pharmacol. 2024 Jan 15;963:176155. doi: 10.1016/j.ejphar.2023.176155. Epub 2023 Oct 31. Eur J Pharmacol. 2024. PMID: 37914065 Review.
-
Sirtuin functions and modulation: from chemistry to the clinic.Clin Epigenetics. 2016 May 25;8:61. doi: 10.1186/s13148-016-0224-3. eCollection 2016. Clin Epigenetics. 2016. PMID: 27226812 Free PMC article. Review.
-
Sirtuin family in autoimmune diseases.Front Immunol. 2023 Jul 6;14:1186231. doi: 10.3389/fimmu.2023.1186231. eCollection 2023. Front Immunol. 2023. PMID: 37483618 Free PMC article. Review.
-
Research progress on Sirtuins (SIRTs) family modulators.Biomed Pharmacother. 2024 May;174:116481. doi: 10.1016/j.biopha.2024.116481. Epub 2024 Mar 24. Biomed Pharmacother. 2024. PMID: 38522239 Review.
Cited by
-
The dual functions of the pentacyclic triterpenoid madecassic acid in ameliorating doxorubicin-induced cardiotoxicity and enhancing the antitumor efficacy of doxorubicin.Int J Biol Sci. 2024 Oct 7;20(14):5396-5414. doi: 10.7150/ijbs.97418. eCollection 2024. Int J Biol Sci. 2024. PMID: 39494326 Free PMC article.
-
Genetic Background of Medication-Related Osteonecrosis of the Jaw: Current Evidence and Future Perspectives.Int J Mol Sci. 2024 Sep 29;25(19):10488. doi: 10.3390/ijms251910488. Int J Mol Sci. 2024. PMID: 39408816 Free PMC article. Review.
-
Monitoring nucleolar-nucleoplasmic protein shuttling in living cells by high-content microscopy and automated image analysis.Nucleic Acids Res. 2024 Aug 27;52(15):e72. doi: 10.1093/nar/gkae598. Nucleic Acids Res. 2024. PMID: 39036969 Free PMC article.
-
The Current State of Research on Sirtuin-Mediated Autophagy in Cardiovascular Diseases.J Cardiovasc Dev Dis. 2023 Sep 6;10(9):382. doi: 10.3390/jcdd10090382. J Cardiovasc Dev Dis. 2023. PMID: 37754811 Free PMC article. Review.
-
Sirtuin 3 ameliorates inflammatory bowel disease via inhibiting intestinal inflammation and oxidative stress.J Clin Biochem Nutr. 2024 May;74(3):235-244. doi: 10.3164/jcbn.23-42. Epub 2024 Feb 9. J Clin Biochem Nutr. 2024. PMID: 38799140 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

