DUSP6 mediates resistance to JAK2 inhibition and drives leukemic progression
- PMID: 36581736
- PMCID: PMC11288645
- DOI: 10.1038/s43018-022-00486-8
DUSP6 mediates resistance to JAK2 inhibition and drives leukemic progression
Abstract
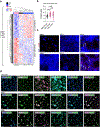
Myeloproliferative neoplasms (MPNs) exhibit a propensity for transformation to secondary acute myeloid leukemia (sAML), for which the underlying mechanisms remain poorly understood, resulting in limited treatment options and dismal clinical outcomes. Here, we performed single-cell RNA sequencing on serial MPN and sAML patient stem and progenitor cells, identifying aberrantly increased expression of DUSP6 underlying disease transformation. Pharmacologic dual-specificity phosphatase (DUSP)6 targeting led to inhibition of S6 and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling while also reducing inflammatory cytokine production. DUSP6 perturbation further inhibited ribosomal S6 kinase (RSK)1, which we identified as a second indispensable candidate associated with poor clinical outcome. Ectopic expression of DUSP6 mediated JAK2-inhibitor resistance and exacerbated disease severity in patient-derived xenograft (PDX) models. Contrastingly, DUSP6 inhibition potently suppressed disease development across Jak2V617F and MPLW515L MPN mouse models and sAML PDXs without inducing toxicity in healthy controls. These findings underscore DUSP6 in driving disease transformation and highlight the DUSP6-RSK1 axis as a vulnerable, druggable pathway in myeloid malignancies.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures






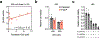










References
-
- Oh ST & Gotlib J. JAK2 V617F and beyond: role of genetics and aberrant signaling in the pathogenesis of myeloproliferative neoplasms. Expert Rev. Hematol 3, 323–337 (2010). - PubMed
-
- Zoi K. & Cross NC Genomics of myeloproliferative neoplasms. J. Clin. Oncol 35, 947–954 (2017). - PubMed
-
- Tefferi A. Myelofibrosis with myeloid metaplasia. N. Engl. J. Med 342, 1255–1265 (2000). - PubMed
-
- Harrison C. et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N. Engl. J. Med 366, 787–798 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

