High-risk genotypes for type 1 diabetes are associated with the imbalance of gut microbiome and serum metabolites
- PMID: 36582242
- PMCID: PMC9794034
- DOI: 10.3389/fimmu.2022.1033393
High-risk genotypes for type 1 diabetes are associated with the imbalance of gut microbiome and serum metabolites
Abstract
Background: The profile of gut microbiota, serum metabolites, and lipids of type 1 diabetes (T1D) patients with different human leukocyte antigen (HLA) genotypes remains unknown. We aimed to explore gut microbiota, serum metabolites, and lipids signatures in individuals with T1D typed by HLA genotypes.
Methods: We did a cross-sectional study that included 73 T1D adult patients. Patients were categorized into two groups according to the HLA haplotypes they carried: those with any two of three susceptibility haplotypes (DR3, DR4, DR9) and without any of the protective haplotypes (DR8, DR11, DR12, DR15, DR16) were defined as high-risk HLA genotypes group (HR, n=30); those with just one or without susceptibility haplotypes as the non-high-risk HLA genotypes group (NHR, n=43). We characterized the gut microbiome profile with 16S rRNA gene amplicon sequencing and analyzed serum metabolites with liquid chromatography-mass spectrometry.
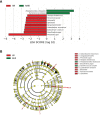
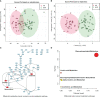
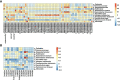
Results: Study individuals were 32.5 (8.18) years old, and 60.3% were female. Compared to NHR, the gut microbiota of HR patients were characterized by elevated abundances of Prevotella copri and lowered abundances of Parabacteroides distasonis. Differential serum metabolites (hypoxanthine, inosine, and guanine) which increased in HR were involved in purine metabolism. Different lipids, phosphatidylcholines and phosphatidylethanolamines, decreased in HR group. Notably, Parabacteroides distasonis was negatively associated (p ≤ 0.01) with hypoxanthine involved in purine metabolic pathways.
Conclusions: The present findings enabled a better understanding of the changes in gut microbiome and serum metabolome in T1D patients with HLA risk genotypes. Alterations of the gut microbiota and serum metabolites may provide some information for distinguishing T1D patients with different HLA risk genotypes.
Keywords: gut microbiota; human leukocyte antigen; serum lipids; serum metabolites; type 1 diabetes mellitus.
Copyright © 2022 Yue, Tan, Wang, Liu, Yang, Ding, Xu, Yan, Zheng, Weng and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
HLA Genetic Discrepancy Between Latent Autoimmune Diabetes in Adults and Type 1 Diabetes: LADA China Study No. 6.J Clin Endocrinol Metab. 2016 Apr;101(4):1693-700. doi: 10.1210/jc.2015-3771. Epub 2016 Feb 11. J Clin Endocrinol Metab. 2016. PMID: 26866570
-
Gut microbiota, serum metabolites, and lipids related to blood glucose control and type 1 diabetes.J Diabetes. 2024 Oct;16(10):e70021. doi: 10.1111/1753-0407.70021. J Diabetes. 2024. PMID: 39463013 Free PMC article.
-
HLA Class II Allele, Haplotype, and Genotype Associations with Type 1 Diabetes in Benin: A Pilot Study.J Diabetes Res. 2017;2017:6053764. doi: 10.1155/2017/6053764. Epub 2017 Jul 20. J Diabetes Res. 2017. PMID: 28808665 Free PMC article.
-
Genetic susceptibility factors of Type 1 diabetes in Asians.Diabetes Metab Res Rev. 2001 Jan-Feb;17(1):2-11. doi: 10.1002/1520-7560(2000)9999:9999<::aid-dmrr164>3.0.co;2-m. Diabetes Metab Res Rev. 2001. PMID: 11241886 Review.
-
Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes.Int J Mol Sci. 2020 Dec 24;22(1):125. doi: 10.3390/ijms22010125. Int J Mol Sci. 2020. PMID: 33374418 Free PMC article. Review.
Cited by
-
A mendelian randomization study investigating the causal relationships between 1400 serum metabolites and autoimmune diseases.Heliyon. 2024 Jul 14;10(14):e34560. doi: 10.1016/j.heliyon.2024.e34560. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39114021 Free PMC article.
-
Host Transcriptome and Microbial Variation in Relation to Visceral Hyperalgesia.Nutrients. 2025 Mar 6;17(5):921. doi: 10.3390/nu17050921. Nutrients. 2025. PMID: 40077792 Free PMC article.
-
The dynamics of the gut microbiota in prediabetes during a four-year follow-up among European patients-an IMI-DIRECT prospective study.Genome Med. 2025 Jul 15;17(1):78. doi: 10.1186/s13073-025-01508-7. Genome Med. 2025. PMID: 40665409 Free PMC article.
-
Important denominator between autoimmune comorbidities: a review of class II HLA, autoimmune disease, and the gut.Front Immunol. 2023 Sep 26;14:1270488. doi: 10.3389/fimmu.2023.1270488. eCollection 2023. Front Immunol. 2023. PMID: 37828987 Free PMC article. Review.
-
Gut microbiome and blood glucose control in type 1 diabetes: a systematic review.Front Endocrinol (Lausanne). 2023 Nov 15;14:1265696. doi: 10.3389/fendo.2023.1265696. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 38034007 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials

