The 'ForensOMICS' approach for postmortem interval estimation from human bone by integrating metabolomics, lipidomics, and proteomics
- PMID: 36583441
- PMCID: PMC9803353
- DOI: 10.7554/eLife.83658
The 'ForensOMICS' approach for postmortem interval estimation from human bone by integrating metabolomics, lipidomics, and proteomics
Abstract
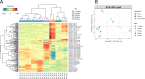
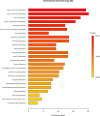
The combined use of multiple omics allows to study complex interrelated biological processes in their entirety. We applied a combination of metabolomics, lipidomics and proteomics to human bones to investigate their combined potential to estimate time elapsed since death (i.e., the postmortem interval [PMI]). This 'ForensOMICS' approach has the potential to improve accuracy and precision of PMI estimation of skeletonized human remains, thereby helping forensic investigators to establish the timeline of events surrounding death. Anterior midshaft tibial bone was collected from four female body donors before their placement at the Forensic Anthropology Research Facility owned by the Forensic Anthropological Center at Texas State (FACTS). Bone samples were again collected at selected PMIs (219-790-834-872days). Liquid chromatography mass spectrometry (LC-MS) was used to obtain untargeted metabolomic, lipidomic, and proteomic profiles from the pre- and post-placement bone samples. The three omics blocks were investigated independently by univariate and multivariate analyses, followed by Data Integration Analysis for Biomarker discovery using Latent variable approaches for Omics studies (DIABLO), to identify the reduced number of markers describing postmortem changes and discriminating the individuals based on their PMI. The resulting model showed that pre-placement metabolome, lipidome and proteome profiles were clearly distinguishable from post-placement ones. Metabolites in the pre-placement samples suggested an extinction of the energetic metabolism and a switch towards another source of fuelling (e.g., structural proteins). We were able to identify certain biomolecules with an excellent potential for PMI estimation, predominantly the biomolecules from the metabolomics block. Our findings suggest that, by targeting a combination of compounds with different postmortem stability, in the future we could be able to estimate both short PMIs, by using metabolites and lipids, and longer PMIs, by using proteins.
Keywords: biochemistry; chemical biology; decomposition; human; human bone; lipidomics; metabolomics; multi-omics; postmortem interval; proteomics.
© 2022, Bonicelli et al.
Conflict of interest statement
AB, HM, AC, EL, DW, NP No competing interests declared
Figures








Update of
- doi: 10.1101/2022.09.29.510059
References
-
- Alldritt I, Whitham-Agut B, Sipin M, Studholme J, Trentacoste A, Tripp JA, Cappai MG, Ditchfield P, Devièse T, Hedges REM, McCullagh JSO. Metabolomics reveals diet-derived plant polyphenols accumulate in physiological bone. Scientific Reports. 2019;9:8047. doi: 10.1038/s41598-019-44390-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

