Identification of potential antigenic peptides of Brucella through proteome and peptidome
- PMID: 36583994
- PMCID: PMC9856995
- DOI: 10.1002/vms3.1048
Identification of potential antigenic peptides of Brucella through proteome and peptidome
Abstract
Background: Brucellosis, caused by Brucella spp., is a major zoonotic public health threat. Although several Brucella vaccines have been demonstrated for use in animals, Brucella spp. can cause human infection and to date, there are no human-use vaccines licensed by any agency. Recently, methods in vaccine informatics have made major breakthroughs in peptide-based epitopes, opening up a new avenue of vaccine development.
Objectives: The purpose of this article was to identify potential antigenic peptides in Brucella by proteome and peptidome analyses.
Methods: Mouse infection models were first established by injection with Brucella and spleen protein profiles were then analysed. Subsequently, the major histocompatibility complex class I or II (major histocompatibility complex [MHC]-I/II)-binding peptides in blood samples were collected by immunoprecipitation and peptides derived from Brucella proteins were identified through liquid chromatography-mass spectrometry (LC-MS/MS). These peptides were then evaluated in a variety of ways, such as in terms of conservation in Brucella and synchronicity in predicted peptides (similarity and coverage), which allowed us to more effectively measure their antigenic potential.
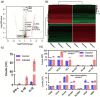
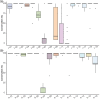
Results: The expression of the inflammatory cytokines IL1B and IFN-γ was significantly altered in the spleen of infected mice and some Brucella proteins, such as Muri, AcpP and GroES, were also detected. Meanwhile, in blood, 35 peptides were identified and most showed high conservation, highlighting their potential as antigen epitopes for vaccine development. In particular, we identified four proteins containing both MHC-I- and MHC-II-binding peptides including AtpA, AtpD, DnaK and BAbS19_II02030. They were also compared with the predicted peptides to estimate their reliability.
Conclusions: The peptides we screened could bind to MHC molecules. After being stimulated with antigen T epitopes, Memory T cells can stimulate T cell activation and promote immune responses. Our results indicated that the peptides we identified may be good candidate targets for the design of subunit vaccines and these results pave the way for the study of safer vaccines against Brucella.
Keywords: Brucella abortus; MHC; mass spectrometry; proteomics.
© 2022 The Authors. Veterinary Medicine and Science published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Prediction of T cell epitopes of Brucella abortus and evaluation of their protective role in mice.Appl Microbiol Biotechnol. 2015 Sep;99(18):7625-37. doi: 10.1007/s00253-015-6787-7. Epub 2015 Jul 7. Appl Microbiol Biotechnol. 2015. PMID: 26150246
-
Identification of MHC-I-Presented Porcine Respiratory and Reproductive Syndrome Virus (PRRSV) Peptides Reveals Immunogenic Epitopes within Several Non-Structural Proteins Recognized by CD8+ T Cells.Viruses. 2022 Aug 26;14(9):1891. doi: 10.3390/v14091891. Viruses. 2022. PMID: 36146698 Free PMC article.
-
Immunoproteomic analysis of Brucella melitensis and identification of a new immunogenic candidate protein for the development of brucellosis subunit vaccine.Mol Immunol. 2011 Oct;49(1-2):175-84. doi: 10.1016/j.molimm.2011.08.009. Epub 2011 Sep 22. Mol Immunol. 2011. PMID: 21943783
-
Progress toward an artificial vaccine for HIV: identification of helper and cytotoxic T-cell epitopes and methods of immunization.Biotechnol Ther. 1991;2(1-2):123-35. Biotechnol Ther. 1991. PMID: 1726961 Review.
-
Mass spectrometry and peptide-based vaccine development.Clin Pharmacol Ther. 2007 Dec;82(6):644-52. doi: 10.1038/sj.clpt.6100389. Epub 2007 Oct 31. Clin Pharmacol Ther. 2007. PMID: 17971823 Review.
Cited by
-
Safe Subunit Green Vaccines Confer Robust Immunity and Protection against Mucosal Brucella Infection in Mice.Vaccines (Basel). 2023 Feb 25;11(3):546. doi: 10.3390/vaccines11030546. Vaccines (Basel). 2023. PMID: 36992130 Free PMC article.
-
Unravelling potential biomarkers for acute and chronic brucellosis through proteomic and bioinformatic approaches.Front Cell Infect Microbiol. 2023 Jul 13;13:1216176. doi: 10.3389/fcimb.2023.1216176. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37520434 Free PMC article.
-
OnmiMHC: a machine learning solution for UCEC tumor vaccine development through enhanced peptide-MHC binding prediction.Front Immunol. 2025 Feb 28;16:1550252. doi: 10.3389/fimmu.2025.1550252. eCollection 2025. Front Immunol. 2025. PMID: 40092998 Free PMC article.
References
-
- Bellacosa, A. (2001). Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ, 8(11), 1076–92. - PubMed
-
- Bourdette, D. , Edmonds, E. , Smith, C. , Bowen, J. D. , Guttmann, C. R. , Nagy, Z. P. , Simon, J. , Whitham, R. , Lovera, J. , Yadav, V. , Mass, M. , Spencer, L. , Culbertson, N. , Bartholomew, R. M. , Theofan, G. , Milano, J. , Offner, H. , & Vandenbark, A. A. (2005). A highly immunogenic trivalent T cell receptor peptide vaccine for multiple sclerosis. Multiple Sclerosis, 11(5), 552–561. 10.1191/1352458505ms1225oa - DOI - PubMed
-
- Buus, S. , Lauemoller, S. L. , Worning, P. , Kesmir, C. , Frimurer, T. , Corbet, S. , Fomsgaard, A. , Hilden, J. , Holm, A. , & Brunak, S. (2003). Sensitive quantitative predictions of peptide‐MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens, 62(5), 378–384. 10.1034/j.1399-0039.2003.00112.x - DOI - PubMed
-
- Chen, Z. , Zhu, Y. , Sha, T. , Zhu, Y. , Sha, T. , Li, Z. , Li, Y. , Zhang, F. , & Ding, J. (2021). Design of a new multi‐epitope vaccine against Brucella based on T and B cell epitopes using bioinformatics methods. Epidemiology and Infection, 149, e136. 10.1017/S0950268821001229 - DOI - PMC - PubMed
-
- Confer, A. W. , Hall, S. M. , Faulkner, C. B. , Espe, B. H. , Deyoe, B. L. , Morton, R. J. , & Smith, R. A. (1985). Effects of challenge dose on the clinical and immune‐responses of cattle vaccinated with reduced doses of Brucella abortus strain‐19. Veterinary Microbiology, 10(6), 561–575. 10.1016/0378-1135(85)90065-3 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

