The inside scoop: Comparative genomics of two intranuclear bacteria, "Candidatus Berkiella cookevillensis" and "Candidatus Berkiella aquae"
- PMID: 36584052
- PMCID: PMC9803151
- DOI: 10.1371/journal.pone.0278206
The inside scoop: Comparative genomics of two intranuclear bacteria, "Candidatus Berkiella cookevillensis" and "Candidatus Berkiella aquae"
Abstract
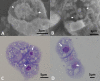
"Candidatus Berkiella cookevillensis" (strain CC99) and "Candidatus Berkiella aquae" (strain HT99), belonging to the Coxiellaceae family, are gram-negative bacteria isolated from amoebae in biofilms present in human-constructed water systems. Both bacteria are obligately intracellular, requiring host cells for growth and replication. The intracellular bacteria-containing vacuoles of both bacteria closely associate with or enter the nuclei of their host cells. In this study, we analyzed the genome sequences of CC99 and HT99 to better understand their biology and intracellular lifestyles. The CC99 genome has a size of 2.9Mb (37.9% GC) and contains 2,651 protein-encoding genes (PEGs) while the HT99 genome has a size of 3.6Mb (39.4% GC) and contains 3,238 PEGs. Both bacteria encode high proportions of hypothetical proteins (CC99: 46.5%; HT99: 51.3%). The central metabolic pathways of both bacteria appear largely intact. Genes for enzymes involved in the glycolytic pathway, the non-oxidative branch of the phosphate pathway, the tricarboxylic acid pathway, and the respiratory chain were present. Both bacteria, however, are missing genes for the synthesis of several amino acids, suggesting reliance on their host for amino acids and intermediates. Genes for type I and type IV (dot/icm) secretion systems as well as type IV pili were identified in both bacteria. Moreover, both bacteria contain genes encoding large numbers of putative effector proteins, including several with eukaryotic-like domains such as, ankyrin repeats, tetratricopeptide repeats, and leucine-rich repeats, characteristic of other intracellular bacteria.
Copyright: © 2022 Kidane et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Description of 'Candidatus Berkiella aquae' and 'Candidatus Berkiella cookevillensis', two intranuclear bacteria of freshwater amoebae.Int J Syst Evol Microbiol. 2016 Feb;66(2):536-541. doi: 10.1099/ijsem.0.000750. Epub 2015 Nov 9. Int J Syst Evol Microbiol. 2016. PMID: 26556637
-
Draft Genome Sequences of Two Novel Amoeba-Resistant Intranuclear Bacteria, "Candidatus Berkiella cookevillensis" and "Candidatus Berkiella aquae".Genome Announc. 2016 Feb 18;4(1):e01732-15. doi: 10.1128/genomeA.01732-15. Genome Announc. 2016. PMID: 26893424 Free PMC article.
-
Infection and nuclear interaction in mammalian cells by 'Candidatus Berkiella cookevillensis', a novel bacterium isolated from amoebae.BMC Microbiol. 2019 May 9;19(1):91. doi: 10.1186/s12866-019-1457-z. BMC Microbiol. 2019. PMID: 31072343 Free PMC article.
-
Coxiella burnetii secretion systems.Adv Exp Med Biol. 2012;984:171-97. doi: 10.1007/978-94-007-4315-1_9. Adv Exp Med Biol. 2012. PMID: 22711632 Review.
-
Rickettsial evolution in the light of comparative genomics.Biol Rev Camb Philos Soc. 2011 May;86(2):379-405. doi: 10.1111/j.1469-185X.2010.00151.x. Epub 2010 Aug 17. Biol Rev Camb Philos Soc. 2011. PMID: 20716256 Review.
Cited by
-
Host-bacteria interactions: ecological and evolutionary insights from ancient, professional endosymbionts.FEMS Microbiol Rev. 2024 Jun 20;48(4):fuae021. doi: 10.1093/femsre/fuae021. FEMS Microbiol Rev. 2024. PMID: 39081075 Free PMC article. Review.
-
Legionella pneumophila type II secretome reveals a polysaccharide deacetylase that impacts intracellular infection, biofilm formation, and resistance to polymyxin- and serum-mediated killing.mBio. 2025 Jul 9;16(7):e0139325. doi: 10.1128/mbio.01393-25. Epub 2025 Jun 20. mBio. 2025. PMID: 40539790 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

