Ancient multiplicity in cyclic nucleotide-gated (CNG) cation channel repertoire was reduced in the ancestor of Olfactores before re-expansion by whole genome duplications in vertebrates
- PMID: 36584110
- PMCID: PMC9803222
- DOI: 10.1371/journal.pone.0279548
Ancient multiplicity in cyclic nucleotide-gated (CNG) cation channel repertoire was reduced in the ancestor of Olfactores before re-expansion by whole genome duplications in vertebrates
Abstract
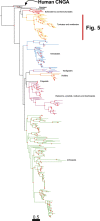
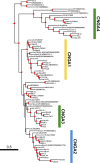
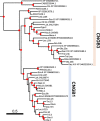
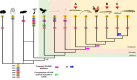
Cyclic nucleotide-gated (CNG) cation channels are important heterotetrameric proteins in the retina, with different subunit composition in cone and rod photoreceptor cells: three CNGA3 and one CNGB3 in cones and three CNGA1 and one CNGB1 in rods. CNGA and CNGB subunits form separate subfamilies. We have analyzed the evolution of the CNG gene family in metazoans, with special focus on vertebrates by using sequence-based phylogeny and conservation of chromosomal synteny to deduce paralogons resulting from the early vertebrate whole genome duplications (WGDs). Our analyses show, unexpectedly, that the CNGA subfamily had four sister subfamilies in the ancestor of bilaterians and cnidarians that we named CNGC, CNGD, CNGE and CNGF. Of these, CNGC, CNGE and CNGF were lost in the ancestor of Olfactores while CNGD was lost in the vertebrate ancestor. The remaining CNGA and CNGB genes were expanded by a local duplication of CNGA and the subsequent chromosome duplications in the basal vertebrate WGD events. Upon some losses, this resulted in the gnathostome ancestor having three members in the visual CNGA subfamily (CNGA1-3), a single CNGA4 gene, and two members in the CNGB subfamily (CNGB1 and CNGB3). The nature of chromosomal rearrangements in the vertebrate CNGA paralogon was resolved by including the genomes of a non-teleost actinopterygian and an elasmobranch. After the teleost-specific WGD, additional duplicates were generated and retained for CNGA1, CNGA2, CNGA3 and CNGB1. Furthermore, teleosts retain a local duplicate of CNGB3. The retention of duplicated CNG genes is explained by their subfunctionalisation and photoreceptor-specific expression. In conclusion, this study provides evidence for four previously unknown CNG subfamilies in metazoans and further evidence that the early vertebrate WGD events were instrumental in the evolution of the vertebrate visual and central nervous systems.
Copyright: © 2022 Lagman et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
The structure of the native CNGA1/CNGB1 CNG channel from bovine retinal rods.Nat Struct Mol Biol. 2022 Jan;29(1):32-39. doi: 10.1038/s41594-021-00700-8. Epub 2021 Dec 30. Nat Struct Mol Biol. 2022. PMID: 34969975
-
The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications.BMC Evol Biol. 2013 Nov 2;13:238. doi: 10.1186/1471-2148-13-238. BMC Evol Biol. 2013. PMID: 24180662 Free PMC article.
-
Native cone photoreceptor cyclic nucleotide-gated channel is a heterotetrameric complex comprising both CNGA3 and CNGB3: a study using the cone-dominant retina of Nrl-/- mice.J Neurochem. 2008 Sep;106(5):2042-55. doi: 10.1111/j.1471-4159.2008.05548.x. Epub 2008 Jul 4. J Neurochem. 2008. PMID: 18665891 Free PMC article.
-
Retinal Cyclic Nucleotide-Gated Channel Regulation by Calmodulin.Int J Mol Sci. 2022 Nov 16;23(22):14143. doi: 10.3390/ijms232214143. Int J Mol Sci. 2022. PMID: 36430626 Free PMC article. Review.
-
Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels.J Biol Chem. 2003 May 23;278(21):18705-8. doi: 10.1074/jbc.R300001200. Epub 2003 Mar 7. J Biol Chem. 2003. PMID: 12626507 Review.
References
-
- Darwin C., Drawin On the origin of species by means of natural selection, 1859.
-
- Lagman D., Daza D.O., Widmark J., Abalo X.M., Sundström G., Larhammar D., The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications, BMC Evol Biol. 13 (2013) 238. doi: 10.1186/1471-2148-13-238 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

