The chaperone protein p32 stabilizes HIV-1 Tat and strengthens the p-TEFb/RNAPII/TAR complex promoting HIV transcription elongation
- PMID: 36584296
- PMCID: PMC9910500
- DOI: 10.1073/pnas.2217476120
The chaperone protein p32 stabilizes HIV-1 Tat and strengthens the p-TEFb/RNAPII/TAR complex promoting HIV transcription elongation
Abstract
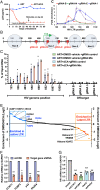
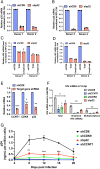
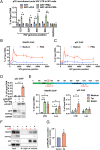
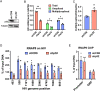
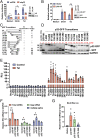
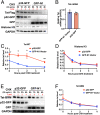
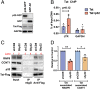
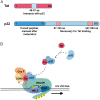
HIV gene expression is modulated by the combinatorial activity of the HIV transcriptional activator, Tat, host transcription factors, and chromatin remodeling complexes. To identify host factors regulating HIV transcription, we used specific single-guide RNAs and endonuclease-deficient Cas9 to perform chromatin affinity purification of the integrated HIV promoter followed by mass spectrometry. The scaffold protein, p32, also called ASF/SF2 splicing factor-associated protein, was identified among the top enriched factors present in actively transcribing HIV promoters but absent in silenced ones. Chromatin immunoprecipitation analysis confirmed the presence of p32 on active HIV promoters and its enhanced recruitment by Tat. HIV uses Tat to efficiently recruit positive transcription elongation factor b (p-TEFb) (CDK9/CCNT1) to TAR, an RNA secondary structure that forms from the first 59 bp of HIV transcripts, to enhance RNAPII transcriptional elongation. The RNA interference of p32 significantly reduced HIV transcription in primary CD4+T cells and in HIV chronically infected cells, independently of either HIV splicing or p32 anti-splicing activity. Conversely, overexpression of p32 specifically increased Tat-dependent HIV transcription. p32 was found to directly interact with Tat's basic domain enhancing Tat stability and half-life. Conversely, p32 associates with Tat via N- and C-terminal domains. Likely due its scaffold properties, p32 also promoted Tat association with TAR, p-TEFb, and RNAPII enhancing Tat-dependent HIV transcription. In sum, we identified p32 as a host factor that interacts with and stabilizes Tat protein, promotes Tat-dependent transcriptional regulation, and may be explored for HIV-targeted transcriptional inhibition.
Keywords: HIV; Tat; p32; regulation; transcription.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat.Genes Dev. 2005 May 15;19(10):1211-26. doi: 10.1101/gad.1291705. Genes Dev. 2005. PMID: 15905409 Free PMC article.
-
CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA.Mol Cell Biol. 2000 Sep;20(18):6958-69. doi: 10.1128/MCB.20.18.6958-6969.2000. Mol Cell Biol. 2000. PMID: 10958691 Free PMC article.
-
Relief of two built-In autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter.Mol Cell Biol. 2000 Aug;20(16):5897-907. doi: 10.1128/MCB.20.16.5897-5907.2000. Mol Cell Biol. 2000. PMID: 10913173 Free PMC article.
-
The control of HIV transcription: keeping RNA polymerase II on track.Cell Host Microbe. 2011 Nov 17;10(5):426-35. doi: 10.1016/j.chom.2011.11.002. Cell Host Microbe. 2011. PMID: 22100159 Free PMC article. Review.
-
Tackling Tat.J Mol Biol. 1999 Oct 22;293(2):235-54. doi: 10.1006/jmbi.1999.3060. J Mol Biol. 1999. PMID: 10550206 Review.
Cited by
-
Breaking the Silence: Regulation of HIV Transcription and Latency on the Road to a Cure.Viruses. 2023 Dec 15;15(12):2435. doi: 10.3390/v15122435. Viruses. 2023. PMID: 38140676 Free PMC article. Review.
-
FUBP3 enhances HIV-1 transcriptional activity and regulates immune response pathways in T cells.Mol Ther Nucleic Acids. 2025 Mar 25;36(2):102525. doi: 10.1016/j.omtn.2025.102525. eCollection 2025 Jun 10. Mol Ther Nucleic Acids. 2025. PMID: 40248217 Free PMC article.
-
HIV-1 transcriptional modulation: novel host factors and prospective therapeutic strategies.Curr Opin HIV AIDS. 2023 Sep 1;18(5):264-272. doi: 10.1097/COH.0000000000000808. Epub 2023 Jul 17. Curr Opin HIV AIDS. 2023. PMID: 37535041 Free PMC article. Review.
-
PRMT3 reverses HIV-1 latency by increasing chromatin accessibility to form a TEAD4-P-TEFb-containing transcriptional hub.Nat Commun. 2025 May 15;16(1):4529. doi: 10.1038/s41467-025-59578-5. Nat Commun. 2025. PMID: 40374607 Free PMC article.
-
Nanoparticle delivery of Tat synergizes with classical latency reversal agents to express HIV antigen targets.Antimicrob Agents Chemother. 2024 Jul 9;68(7):e0020124. doi: 10.1128/aac.00201-24. Epub 2024 Jun 3. Antimicrob Agents Chemother. 2024. PMID: 38829049 Free PMC article.
References
-
- Chun T. W., et al. , Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 197, 714–720 (2008). - PubMed
-
- Gunthard H. F., et al. , Residual human immunodeficiency virus (HIV) Type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years. J. Infect. Dis. 183, 1318–1327 (2001). - PubMed
-
- Knuchel M. C., et al. , Analysis of a biallelic polymorphism in the tumor necrosis factor alpha promoter and HIV type 1 disease progression. AIDS Res. Hum. Retroviruses 14, 305–309 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

