Unusual phototransduction via cross-motif signaling from Gq to adenylyl cyclase in intrinsically photosensitive retinalganglion cells
- PMID: 36584299
- PMCID: PMC9910442
- DOI: 10.1073/pnas.2216599120
Unusual phototransduction via cross-motif signaling from Gq to adenylyl cyclase in intrinsically photosensitive retinalganglion cells
Abstract
Nonimage-forming vision in mammals is mediated primarily by melanopsin (OPN4)-expressing, intrinsically photosensitive retinal ganglion cells (ipRGCs). In mouse M1-ipRGCs, melanopsin predominantly activates, via Gαq,11,14, phospholipase C-β4 to open transient receptor 6 (TRPC6) and TRPC7 channels. In M2- and M4-ipRGCs, however, a prominent phototransduction mechanism involves the opening of hyperpolarization- and cyclic nucleotide-gated channels via cyclic nucleotide, although the upstream steps remain uncertain. We report here experiments, primarily on M4-ipRGCs, with photo-uncaging of cyclic nucleotides and virally expressed CNGA2 channels to conclude that the second messenger is cyclic adenosine monophosphate (cAMP) - very surprising considering that cyclic guanosine monophosphate (cGMP) is used in almost all cyclic nucleotide-mediated phototransduction mechanisms across the animal kingdom. We further found that the upstream G protein is likewise Gq, which via its Gβγ subunits directly activates adenylyl cyclase (AC). Our findings are a demonstration in a native cell of a cross-motif GPCR signaling pathway from Gq directly to AC with a specific function.
Keywords: Gq; cAMP; ipRGC; melanopsin; phototransduction.
Conflict of interest statement
The authors declare no competing interest.
Figures


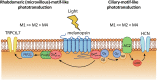
Similar articles
-
Cyclic-Nucleotide- and HCN-Channel-Mediated Phototransduction in Intrinsically Photosensitive Retinal Ganglion Cells.Cell. 2018 Oct 18;175(3):652-664.e12. doi: 10.1016/j.cell.2018.08.055. Epub 2018 Sep 27. Cell. 2018. PMID: 30270038 Free PMC article.
-
Loss of gq/11 genes does not abolish melanopsin phototransduction.PLoS One. 2014 May 28;9(5):e98356. doi: 10.1371/journal.pone.0098356. eCollection 2014. PLoS One. 2014. PMID: 24870805 Free PMC article.
-
Intrinsic phototransduction persists in melanopsin-expressing ganglion cells lacking diacylglycerol-sensitive TRPC subunits.Eur J Neurosci. 2011 Mar;33(5):856-67. doi: 10.1111/j.1460-9568.2010.07583.x. Epub 2011 Jan 24. Eur J Neurosci. 2011. PMID: 21261756 Free PMC article.
-
[Phototransduction mediated by melanopsin in intrinsically photosensitive retinal ganglion cells].Gac Med Mex. 2015 Nov-Dec;151(6):764-76. Gac Med Mex. 2015. PMID: 26581535 Review. Spanish.
-
Melanopsin and mechanisms of non-visual ocular photoreception.J Biol Chem. 2012 Jan 13;287(3):1649-56. doi: 10.1074/jbc.R111.301226. Epub 2011 Nov 10. J Biol Chem. 2012. PMID: 22074930 Free PMC article. Review.
Cited by
-
Comprehensive bibliometric research in neuroscience: focusing on ophthalmology.Front Neurosci. 2023 Jun 15;17:1106023. doi: 10.3389/fnins.2023.1106023. eCollection 2023. Front Neurosci. 2023. PMID: 37397445 Free PMC article.
-
Coexistence within one cell of microvillous and ciliary phototransductions across M1- through M6-IpRGCs.Proc Natl Acad Sci U S A. 2023 Dec 26;120(52):e2315282120. doi: 10.1073/pnas.2315282120. Epub 2023 Dec 18. Proc Natl Acad Sci U S A. 2023. PMID: 38109525 Free PMC article.
-
Tumor metabolic crosstalk and immunotherapy.Clin Transl Oncol. 2024 Apr;26(4):797-807. doi: 10.1007/s12094-023-03304-4. Epub 2023 Sep 23. Clin Transl Oncol. 2024. PMID: 37740892 Review.
-
Melanopsin-mediated optical entrainment regulates circadian rhythms in vertebrates.Commun Biol. 2023 Oct 18;6(1):1054. doi: 10.1038/s42003-023-05432-7. Commun Biol. 2023. PMID: 37853054 Free PMC article. Review.
-
The Role of Retinal Dysfunction in Myopia Development.Cell Mol Neurobiol. 2023 Jul;43(5):1905-1930. doi: 10.1007/s10571-022-01309-1. Epub 2022 Nov 24. Cell Mol Neurobiol. 2023. PMID: 36427109 Free PMC article. Review.
References
-
- Berson D. M., Dunn F. A., Takao M., Phototransduction by retinal ganglion cells that set the circadian clock. Science 295, 1070–1073 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

