Distinct fibroblast progenitor subpopulation expedites regenerative mucosal healing by immunomodulation
- PMID: 36584405
- PMCID: PMC9827523
- DOI: 10.1084/jem.20221350
Distinct fibroblast progenitor subpopulation expedites regenerative mucosal healing by immunomodulation
Abstract
Injuries that heal by fibrosis can compromise organ function and increase patient morbidity. The oral mucosal barrier has a high regenerative capacity with minimal scarring, but the cellular mechanisms remain elusive. Here, we identify distinct postnatal paired-related homeobox-1+ (Prx1+) cells as a critical fibroblast subpopulation that expedites mucosal healing by facilitating early immune response. Using transplantation and genetic ablation model in mice, we show that oral mucosa enriched with Prx1+ cells heals faster than those that lack Prx1+ cells. Lineage tracing and scRNA-seq reveal that Prx1+ fibroblasts exhibit progenitor signatures in physiologic and injured conditions. Mechanistically, Prx1+ progenitors accelerate wound healing by differentiating into immunomodulatory SCA1+ fibroblasts, which prime macrophage recruitment through CCL2 as a key part of pro-wound healing response. Furthermore, human Prx1+ fibroblasts share similar gene and spatial profiles compared to their murine counterpart. Thus, our data suggest that Prx1+ fibroblasts may provide a valuable source in regenerative procedures for the treatment of corneal wounds and enteropathic fibrosis.
© 2022 Ko et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures

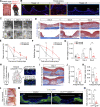
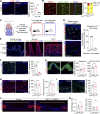




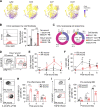


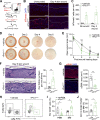


Comment in
-
Fast and scarless: Prx1+ fibroblasts turbocharge healing.J Exp Med. 2023 Mar 6;220(3):e20222139. doi: 10.1084/jem.20222139. Epub 2023 Feb 6. J Exp Med. 2023. PMID: 36745188 Free PMC article.
References
-
- Boniakowski, A.E., Kimball A.S., Joshi A., Schaller M., Davis F.M., denDekker A., Obi A.T., Moore B.B., Kunkel S.L., and Gallagher K.A.. 2018. Murine macrophage chemokine receptor CCR2 plays a crucial role in macrophage recruitment and regulated inflammation in wound healing. Eur. J. Immunol. 48:1445–1455. 10.1002/eji.201747400 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

