Regulation of telomere silencing by the core histones-autophagy-Sir2 axis
- PMID: 36585257
- PMCID: PMC9806677
- DOI: 10.26508/lsa.202201614
Regulation of telomere silencing by the core histones-autophagy-Sir2 axis
Abstract
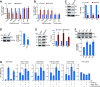
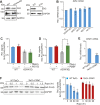
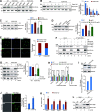
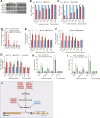
Telomeres contain compacted heterochromatin, and genes adjacent to telomeres are subjected to transcription silencing. Maintaining telomere structure integrity and transcription silencing is important to prevent the occurrence of premature aging and aging-related diseases. How telomere silencing is regulated during aging is not well understood. Here, we find that the four core histones are reduced during yeast chronological aging, leading to compromised telomere silencing. Mechanistically, histone loss promotes the nuclear export of Sir2 and its degradation by autophagy. Meanwhile, reducing core histones enhances the autophagy pathway, which further accelerates autophagy-mediated Sir2 degradation. By screening the histone mutant library, we identify eight histone mutants and one histone modification (histone methyltransferase Set1-catalyzed H3K4 trimethylation) that regulate telomere silencing by modulating the core histones-autophagy-Sir2 axis. Overall, our findings reveal core histones and autophagy as causes of aging-coupled loss of telomere silencing and shed light on dynamic regulation of telomere structure during aging.
© 2022 Mei et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
-
- Bruhn C, Ajazi A, Ferrari E, Lanz MC, Batrin R, Choudhary R, Walvekar A, Laxman S, Longhese MP, Fabre E, et al. (2020) The Rad53CHK1/CHK2-Spt21NPAT and Tel1ATM axes couple glucose tolerance to histone dosage and subtelomeric silencing. Nat Commun 11: 4154. 10.1038/s41467-020-17961-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
