Expression profiles of small non-coding RNAs in breast cancer tumors characterize clinicopathological features and show prognostic and predictive potential
- PMID: 36585466
- PMCID: PMC9803687
- DOI: 10.1038/s41598-022-26954-w
Expression profiles of small non-coding RNAs in breast cancer tumors characterize clinicopathological features and show prognostic and predictive potential
Abstract
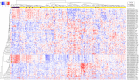
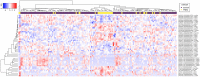
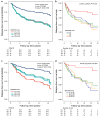
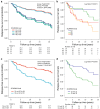
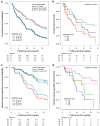
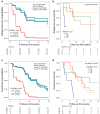
Precision medicine approaches are required for more effective therapies for cancer. As small non-coding RNAs (sncRNAs) have recently been suggested as intriguing candidates for cancer biomarkers and have shown potential also as novel therapeutic targets, we aimed at profiling the non-miRNA sncRNAs in a large sample set to evaluate their role in invasive breast cancer (BC). We used small RNA sequencing and 195 fresh-frozen invasive BC and 22 benign breast tissue samples to identify significant associations of small nucleolar RNAs, small nuclear RNAs, and miscellaneous RNAs with the clinicopathological features and patient outcome of BC. Ninety-six and five sncRNAs significantly distinguished (Padj < 0.01) invasive local BC from benign breast tissue and metastasized BC from invasive local BC, respectively. Furthermore, 69 sncRNAs significantly associated (Padj < 0.01) with the tumor grade, hormone receptor status, subtype, and/or tumor histology. Additionally, 42 sncRNAs were observed as candidates for prognostic markers and 29 for predictive markers for radiotherapy and/or tamoxifen response (P < 0.05). We discovered the clinical relevance of sncRNAs from each studied RNA type. By introducing new sncRNA biomarker candidates for invasive BC and validating the potential of previously described ones, we have guided the way for further research that is warranted for providing novel insights into BC biology.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Small Non-Coding RNAs and Their Role in Locoregional Metastasis and Outcomes in Early-Stage Breast Cancer Patients.Int J Mol Sci. 2024 Apr 3;25(7):3982. doi: 10.3390/ijms25073982. Int J Mol Sci. 2024. PMID: 38612790 Free PMC article.
-
The debatable presence of PIWI-interacting RNAs in invasive breast cancer.Cancer Med. 2021 Jun;10(11):3593-3603. doi: 10.1002/cam4.3915. Epub 2021 May 7. Cancer Med. 2021. PMID: 33960684 Free PMC article.
-
Small non-coding RNAs in human cancer: function, clinical utility, and characterization.Oncogene. 2021 Mar;40(9):1570-1577. doi: 10.1038/s41388-020-01630-3. Epub 2021 Jan 15. Oncogene. 2021. PMID: 33452456 Review.
-
Links between the oncoprotein YB-1 and small non-coding RNAs in breast cancer.PLoS One. 2013 Nov 18;8(11):e80171. doi: 10.1371/journal.pone.0080171. eCollection 2013. PLoS One. 2013. PMID: 24260353 Free PMC article.
-
Exploring the expanding universe of small RNAs.Nat Cell Biol. 2022 Apr;24(4):415-423. doi: 10.1038/s41556-022-00880-5. Epub 2022 Apr 12. Nat Cell Biol. 2022. PMID: 35414016 Free PMC article. Review.
Cited by
-
Combination of multiple omics techniques for a personalized therapy or treatment selection.Front Immunol. 2023 Sep 27;14:1258013. doi: 10.3389/fimmu.2023.1258013. eCollection 2023. Front Immunol. 2023. PMID: 37828984 Free PMC article. Review.
-
HTLV-1 and HTLV-2 infections significantly alter small RNA expression in asymptomatic carriers.Front Med (Lausanne). 2025 Feb 17;12:1547712. doi: 10.3389/fmed.2025.1547712. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40034383 Free PMC article.
-
Small Non-Coding RNAs and Their Role in Locoregional Metastasis and Outcomes in Early-Stage Breast Cancer Patients.Int J Mol Sci. 2024 Apr 3;25(7):3982. doi: 10.3390/ijms25073982. Int J Mol Sci. 2024. PMID: 38612790 Free PMC article.
-
Snord67 promotes breast cancer metastasis by guiding U6 modification and modulating the splicing landscape.Nat Commun. 2025 May 2;16(1):4118. doi: 10.1038/s41467-025-59406-w. Nat Commun. 2025. PMID: 40316533 Free PMC article.
-
Human Vault RNAs: Exploring Their Potential Role in Cellular Metabolism.Int J Mol Sci. 2024 Apr 6;25(7):4072. doi: 10.3390/ijms25074072. Int J Mol Sci. 2024. PMID: 38612882 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

