In Silico and In Vitro Analyses Reveal Promising Antimicrobial Peptides from Myxobacteria
- PMID: 36586039
- PMCID: PMC9839799
- DOI: 10.1007/s12602-022-10036-4
In Silico and In Vitro Analyses Reveal Promising Antimicrobial Peptides from Myxobacteria
Abstract
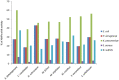
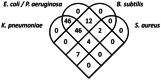
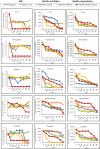
Antimicrobial resistance (AMR) is a global concern, and as soon as new antibiotics are introduced, resistance to those agents emerges. Therefore, there is an increased appetite for alternative antimicrobial agents to traditional antibiotics. Here, we used in silico methods to investigate potential antimicrobial peptides (AMPs) from predatory myxobacteria. Six hundred seventy-two potential AMP sequences were extracted from eight complete myxobacterial genomes. Most putative AMPs were predicted to be active against Klebsiella pneumoniae with least activity being predicted against Staphylococcus aureus. One hundred seventeen AMPs (defined here as 'potent putative AMPs') were predicted to have very good activity against more than two bacterial pathogens, and these were characterized further in silico. All potent putative AMPs were predicted to have anti-inflammatory and antifungal properties, but none was predicted to be active against viruses. Twenty six (22%) of them were predicted to be hemolytic to human erythrocytes, five were predicted to have anticancer properties, and 56 (47%) were predicted to be biofilm active. In vitro assays using four synthesized AMPs showed high MIC values (e.g. So_ce_56_913 250 µg/ml and Coral_AMP411 125 µg/ml against E. coli). However, antibiofilm assays showed a substantial reduction in numbers (e.g. Coral_AMP411 and Myxo_mac104 showed a 69% and 73% reduction, respectively, at the lowest concentration against E. coli) compared to traditional antibiotics. Fourteen putative AMPs had high sequence similarity to proteins which were functionally associated with proteins of known function. The myxobacterial genomes also possessed a variety of biosynthetic gene clusters (BGCs) that can encode antimicrobial secondary metabolites, but their numbers did not correlate with those of the AMPs. We suggest that AMPs from myxobacteria are a promising source of novel antimicrobial agents with a plethora of biological properties.
Keywords: Antimicrobial peptides; Biosynthetic gene clusters; In silico analysis; Myxobacteria.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
In Vitro Evaluation of Antimicrobial Peptides from the Black Soldier Fly (Hermetia Illucens) against a Selection of Human Pathogens.Microbiol Spectr. 2022 Feb 23;10(1):e0166421. doi: 10.1128/spectrum.01664-21. Epub 2022 Jan 5. Microbiol Spectr. 2022. PMID: 34985302 Free PMC article.
-
Aggregation-prone antimicrobial peptides target gram-negative bacterial nucleic acids and protein synthesis.Acta Biomater. 2025 Jan 15;192:446-460. doi: 10.1016/j.actbio.2024.12.002. Epub 2024 Dec 3. Acta Biomater. 2025. PMID: 39637960
-
Novel antimicrobial peptides identified in legume plant, Medicago truncatula.Microbiol Spectr. 2024 Feb 6;12(2):e0182723. doi: 10.1128/spectrum.01827-23. Epub 2024 Jan 18. Microbiol Spectr. 2024. PMID: 38236024 Free PMC article.
-
Beyond Conventional Drug Design: Exploring the Broad-Spectrum Efficacy of Antimicrobial Peptides.Chem Biodivers. 2025 Mar;22(3):e202401349. doi: 10.1002/cbdv.202401349. Epub 2024 Dec 4. Chem Biodivers. 2025. PMID: 39480053 Review.
-
A comprehensive guide on screening and selection of a suitable AMP against biofilm-forming bacteria.Crit Rev Microbiol. 2024 Sep;50(5):859-878. doi: 10.1080/1040841X.2023.2293019. Epub 2023 Dec 15. Crit Rev Microbiol. 2024. PMID: 38102871 Review.
Cited by
-
Design and Synthesis of Novel Antimicrobial Agents.Antibiotics (Basel). 2023 Mar 22;12(3):628. doi: 10.3390/antibiotics12030628. Antibiotics (Basel). 2023. PMID: 36978495 Free PMC article. Review.
-
Machine learning assisted rational design of antimicrobial peptides based on human endogenous proteins and their applications for cosmetic preservative system optimization.Sci Rep. 2024 Jan 10;14(1):947. doi: 10.1038/s41598-023-50832-8. Sci Rep. 2024. PMID: 38200054 Free PMC article.
-
MyxoPortal: a database of myxobacterial genomic features.Database (Oxford). 2024 Jul 2;2024:baae056. doi: 10.1093/database/baae056. Database (Oxford). 2024. PMID: 38958433 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

