Deleterious, protein-altering variants in the transcriptional coregulator ZMYM3 in 27 individuals with a neurodevelopmental delay phenotype
- PMID: 36586412
- PMCID: PMC9943726
- DOI: 10.1016/j.ajhg.2022.12.007
Deleterious, protein-altering variants in the transcriptional coregulator ZMYM3 in 27 individuals with a neurodevelopmental delay phenotype
Abstract
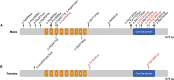
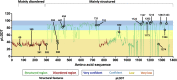
Neurodevelopmental disorders (NDDs) result from highly penetrant variation in hundreds of different genes, some of which have not yet been identified. Using the MatchMaker Exchange, we assembled a cohort of 27 individuals with rare, protein-altering variation in the transcriptional coregulator ZMYM3, located on the X chromosome. Most (n = 24) individuals were males, 17 of which have a maternally inherited variant; six individuals (4 male, 2 female) harbor de novo variants. Overlapping features included developmental delay, intellectual disability, behavioral abnormalities, and a specific facial gestalt in a subset of males. Variants in almost all individuals (n = 26) are missense, including six that recurrently affect two residues. Four unrelated probands were identified with inherited variation affecting Arg441, a site at which variation has been previously seen in NDD-affected siblings, and two individuals have de novo variation resulting in p.Arg1294Cys (c.3880C>T). All variants affect evolutionarily conserved sites, and most are predicted to damage protein structure or function. ZMYM3 is relatively intolerant to variation in the general population, is widely expressed across human tissues, and encodes a component of the KDM1A-RCOR1 chromatin-modifying complex. ChIP-seq experiments on one variant, p.Arg1274Trp, indicate dramatically reduced genomic occupancy, supporting a hypomorphic effect. While we are unable to perform statistical evaluations to definitively support a causative role for variation in ZMYM3, the totality of the evidence, including 27 affected individuals, recurrent variation at two codons, overlapping phenotypic features, protein-modeling data, evolutionary constraint, and experimentally confirmed functional effects strongly support ZMYM3 as an NDD-associated gene.
Keywords: X-linked intellectual disability; ZMYM3; chromatin modifiers; neurodevelopmental disorder; transcriptional coregulators.
Copyright © 2022 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.L.B., Y.C., B.R.L., M.P.N., A.G.N., and H.Z.E. are employees of GeneDx, LLC. S.E.A. is a cofounder and CEO of MediGenome, the Swiss Institute of Genomic Medicine. All other authors declare no competing interests.
Figures


References
-
- Srivastava S., Love-Nichols J.A., Dies K.A., Ledbetter D.H., Martin C.L., Chung W.K., Firth H.V., Frazier T., Hansen R.L., Prock L., et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 2019;21:2413–2421. doi: 10.1038/S41436-019-0554-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

