Flavivirus proteases: The viral Achilles heel to prevent future pandemics
- PMID: 36586467
- PMCID: PMC10062209
- DOI: 10.1016/j.antiviral.2022.105516
Flavivirus proteases: The viral Achilles heel to prevent future pandemics
Abstract
Flaviviruses are important human pathogens and include dengue (DENV), West Nile (WNV), Yellow fever virus (YFV), Japanese encephalitis (JEV) and Zika virus (ZIKV). DENV, transmitted by mosquitoes, causes diseases ranging in severity from mild dengue fever with non-specific flu-like symptoms to fatal dengue hemorrhagic fever and dengue shock syndrome. DENV infections are caused by four serotypes, DENV1-4, which interact differently with antibodies in blood serum. The incidence of DENV infection has increased dramatically in recent decades and the CDC estimates 400 million dengue infections occur each year, resulting in ∼25,000 deaths mostly among children and elderly people. Similarly, ZIKV infections are caused by infected mosquito bites to humans, can be transmitted sexually and through blood transfusions. If a pregnant woman is infected, the virus can cross the placental barrier and can spread to her fetus, causing severe brain malformations in the child including microcephaly and other birth defects. It is noteworthy that the neurological manifestations of ZIKV were also observed in DENV endemic regions, suggesting that pre-existing antibody response to DENV could augment ZIKV infection. WNV, previously unknown in the US (and known to cause only mild disease in Middle East), first arrived in New York city in 1999 (NY99) and spread throughout the US and Canada by Culex mosquitoes and birds. WNV is now endemic in North America. Thus, emerging and re-emerging flaviviruses are significant threat to human health. However, vaccines are available for only a limited number of flaviviruses, and antiviral therapies are not available for any flavivirus. Hence, there is an urgent need to develop therapeutics that interfere with essential enzymatic steps, such as protease in the flavivirus lifecycle as these viruses possess significant threat to future pandemics. In this review, we focus on our E. coli expression of NS2B hydrophilic domain (NS2BH) covalently linked to NS3 protease domain (NS3Pro) in their natural context which is processed by the combined action of both subunits of the NS2B-NS3Pro precursor. Biochemical activities of the viral protease such as solubility and autoproteolysis of NS2BH-NS3Pro linkage depended on the C-terminal portion of NS2BH linked to the NS3Pro domain. Since 2008, we also focus on the use of the recombinant protease in high throughput screens and characterization of small molecular compounds identified in these screens.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
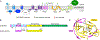

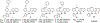
References
-
- Abrams RPM, Yasgar A, Teramoto T, Lee MH, Dorjsuren D, Eastman RT, Malik N, Zakharov AV, Li W, Bachani M, Brimacombe K, Steiner JP, Hall MD, Balasubramanian A, Jadhav A, Padmanabhan R, Simeonov A, Nath A, 2020. Therapeutic candidates for the Zika virus identified by a high-throughput screen for Zika protease inhibitors. Proc Natl Acad Sci U S A 117, 31365–31375. - PMC - PubMed
-
- Akaberi D, Bahlstrom A, Chinthakindi PK, Nyman T, Sandstrom A, Jarhult JD, Palanisamy N, Lundkvist A, Lennerstrand J, 2021. Targeting the NS2B-NS3 protease of tick-borne encephalitis virus with pan-flaviviral protease inhibitors. Antiviral Res 190, 105074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

