Identification and analysis of odorant receptors expressed in the two main olfactory organs, antennae and palps, of Schistocerca americana
- PMID: 36587060
- PMCID: PMC9805433
- DOI: 10.1038/s41598-022-27199-3
Identification and analysis of odorant receptors expressed in the two main olfactory organs, antennae and palps, of Schistocerca americana
Abstract
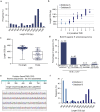
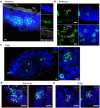
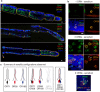
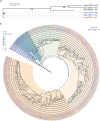
Locusts depend upon their sense of smell and provide useful models for understanding olfaction. Extending this understanding requires knowledge of the molecular and structural organization of the olfactory system. Odor sensing begins with olfactory receptor neurons (ORNs), which express odorant receptors (ORs). In insects, ORNs are housed, in varying numbers, in olfactory sensilla. Because the organization of ORs within sensilla affects their function, it is essential to identify the ORs they contain. Here, using RNA sequencing, we identified 179 putative ORs in the transcriptomes of the two main olfactory organs, antenna and palp, of the locust Schistocerca americana. Quantitative expression analysis showed most putative ORs (140) are expressed in antennae while only 31 are in the palps. Further, our analysis identified one OR detected only in the palps and seven ORs that are expressed differentially by sex. An in situ analysis of OR expression suggested ORs are organized in non-random combinations within antennal sensilla. A phylogenetic comparison of OR predicted protein sequences revealed homologous relationships among two other Acrididae species. Our results provide a foundation for understanding the organization of the first stage of the olfactory system in S. americana, a well-studied model for olfactory processing.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Differential expression of two novel odorant receptors in the locust (Locusta migratoria).BMC Neurosci. 2013 Apr 22;14:50. doi: 10.1186/1471-2202-14-50. BMC Neurosci. 2013. PMID: 23607307 Free PMC article.
-
SNMP1 and odorant receptors are co-expressed in olfactory neurons of the labial and maxillary palps from the desert locust Schistocerca gregaria (Orthoptera: Acrididae).Cell Tissue Res. 2020 Feb;379(2):275-289. doi: 10.1007/s00441-019-03083-x. Epub 2019 Aug 19. Cell Tissue Res. 2020. PMID: 31478139
-
In Search for Pheromone Receptors: Certain Members of the Odorant Receptor Family in the Desert Locust Schistocerca gregaria (Orthoptera: Acrididae) Are Co-expressed with SNMP1.Int J Biol Sci. 2017 Jul 15;13(7):911-922. doi: 10.7150/ijbs.18402. eCollection 2017. Int J Biol Sci. 2017. PMID: 28808423 Free PMC article.
-
Elements of olfactory reception in adult Drosophila melanogaster.Anat Rec (Hoboken). 2013 Sep;296(9):1477-88. doi: 10.1002/ar.22747. Epub 2013 Jul 31. Anat Rec (Hoboken). 2013. PMID: 23904114 Review.
-
Odor Coding in Insects.In: Menini A, editor. The Neurobiology of Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 2. In: Menini A, editor. The Neurobiology of Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 2. PMID: 21882428 Free Books & Documents. Review.
Cited by
-
The conserved IR75 subfamily mediates carboxylic acid detection in insects of public health and agricultural importance.J Insect Sci. 2025 Jan 20;25(1):10. doi: 10.1093/jisesa/ieaf012. J Insect Sci. 2025. PMID: 39891408 Free PMC article. Review.
-
From bristle to brain: embryonic development of topographic projections from basiconic sensilla in the antennal nervous system of the locust Schistocerca gregaria.Dev Genes Evol. 2024 Jun;234(1):33-44. doi: 10.1007/s00427-024-00716-2. Epub 2024 May 1. Dev Genes Evol. 2024. PMID: 38691194 Free PMC article.
References
-
- Murlis J, Willis MA, Carde RT. Spatial and temporal structures of pheromone plumes in fields and forests. Physiol. Entomol. 2000;25(3):211–222. doi: 10.1046/j.1365-3032.2000.00176.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

