Mechanism of autophagy induced by activation of the AMPK/ERK/mTOR signaling pathway after TRIM22-mediated DENV-2 infection of HUVECs
- PMID: 36587218
- PMCID: PMC9805691
- DOI: 10.1186/s12985-022-01932-w
Mechanism of autophagy induced by activation of the AMPK/ERK/mTOR signaling pathway after TRIM22-mediated DENV-2 infection of HUVECs
Abstract
Background: Dengue virus type 2 (DENV-2) was used to infect primary human umbilical vein endothelial cells (HUVECs) to examine autophagy induced by activation of the adenosine monophosphate-activated protein kinase (AMPK)/extracellular signal-regulated kinase (ERK)/mammalian target of rapamycin (mTOR) signaling pathway following tripartite motif-containing 22 (TRIM22)-mediated DENV-2 infection to further reveal the underlying pathogenic mechanism of DENV-2 infection.
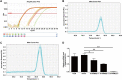
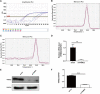
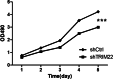
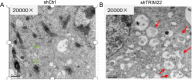
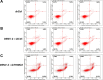
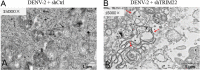
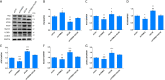
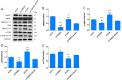
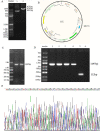
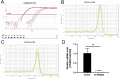
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was used to screen putative interference targets of TRIM22 and determine the knockdown efficiency. The effect of TRIM22 knockdown on HUVEC proliferation was determined using the CCK8 assay. Following TRIM22 knockdown, transmission electron microscopy (TEM) was used to determine the ultrastructure of HUVEC autophagosomes and expression of HUVEC autophagy and AMPK pathway-related genes were measured by qRT-PCR. Moreover, HUVEC autophagy and AMPK pathway-related protein expression levels were determined by western blot analysis. Cell cycle and apoptosis were assessed by flow cytometry (FCM) and the autophagosome structure of the HUVECs was observed by TEM.
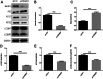
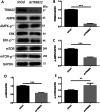
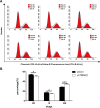
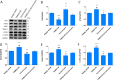
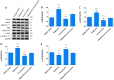
Results: Western blot results indicated that TRIM22 protein expression levels increased significantly 36 h after DENV-2 infection, which was consistent with the proteomics prediction. The CCK8 assay revealed that HUVEC proliferation was reduced following TRIM22 knockdown (P < 0.001). The TEM results indicated that HUVEC autolysosomes increased and autophagy was inhibited after TRIM22 knockdown. The qRT-PCR results revealed that after TRIM22 knockdown, the expression levels of antithymocyte globulin 7 (ATG7), antithymocyte globulin 5 (ATG5), Beclin1, ERK, and mTOR genes decreased (P < 0.01); however, the expression of AMPK genes (P < 0.05) and P62 genes (P < 0.001) increased. FCM revealed that following TRIM22 knockdown, the percentage of HUVECs in the G2 phase increased (P < 0.001) along with cell apoptosis. The effect of TRIM22 overexpression on HUVEC autophagy induced by DENV-2 infection and AMPK pathways decreased after adding an autophagy inhibitor.
Conclusions: In HUVECs, TRIM22 protein positively regulates autophagy and may affect autophagy through the AMPK/ERK/mTOR signaling pathway. Autophagy is induced by activation of the AMPK/ERK/mTOR signaling pathway following TRIM22-mediated DENV-2 infection of HUVECs.
Keywords: AMPK/ERK/mTOR signaling pathway; Autophagy; DENV-2; TRIM22.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


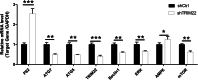
References
-
- Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ, Hay SI, Bedi N, Bensenor IM, Castañeda-Orjuela CA, et al. The global burden of dengue: an analysis from the global burden of disease study 2013. Lancet Infect Dis. 2016;16:712–723. doi: 10.1016/S1473-3099(16)00026-8. - DOI - PMC - PubMed
-
- Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, Kurscheid S, Wang P, Krishnan MN, Higgs S, Fikrig E. Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 2011;7:e1002189. doi: 10.1371/journal.ppat.1002189. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

