Single-cell and spatial analyses reveal the association between gene expression of glutamine synthetase with the immunosuppressive phenotype of APOE+CTSZ+TAM in cancers
- PMID: 36587392
- PMCID: PMC10061288
- DOI: 10.1002/1878-0261.13373
Single-cell and spatial analyses reveal the association between gene expression of glutamine synthetase with the immunosuppressive phenotype of APOE+CTSZ+TAM in cancers
Abstract
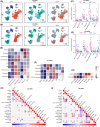
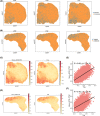
An immunosuppressive state is regulated by various factors in the tumor microenvironment (TME), including, but not limited to, metabolic plasticity of immunosuppressive cells and cytokines secreted by these cells. We used single-cell RNA-sequencing (scRNA-seq) data and applied single-cell flux estimation analysis to characterize the link between metabolism and cellular function within the hypoxic TME of colorectal (CRC) and lung cancer. In terms of metabolic heterogeneity, we found myeloid cells potentially inclined to accumulate glutamine but tumor cells inclined to accumulate glutamate. In particular, we uncovered a tumor-associated macrophage (TAM) subpopulation, APOE+CTSZ+TAM, that was present in high proportions in tumor samples and exhibited immunosuppressive characteristics through upregulating the expression of anti-inflammatory genes. The proportion of APOE+CTSZ+TAM and regulatory T cells (Treg) were positively correlated across CRC scRNA-seq samples. APOE+CTSZ+TAM potentially interacted with Treg via CXCL16-CCR6 signals, as seen by ligand-receptor interactions analysis. Notably, glutamate-to-glutamine metabolic flux score and glutamine synthetase (GLUL) expression were uniquely higher in APOE+CTSZ+TAM, compared with other cell types within the TME. GLUL expression in macrophages was positively correlated with anti-inflammatory score and was higher in high-grade and invasive tumor samples. Moreover, spatial transcriptome and multiplex immunofluorescence staining of samples showed that APOE+CTSZ+TAM and Treg potentially colocalized in the tissue sections from CRC clinical samples. These results highlight the specific role and metabolic characteristic of the APOE+CTSZ+TAM subpopulation and provide a new perspective for macrophage subcluster-targeted therapeutic interventions or metabolic checkpoint-based cancer therapies.
Keywords: GLUL; colorectal cancer; immunosuppressive microenvironment; metabolism; single-cell and spatial RNA-seq; tumor-associated macrophages.
© 2023 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

