Genome-wide identification of Reverse Transcriptase domains of recently inserted endogenous plant pararetrovirus (Caulimoviridae)
- PMID: 36589050
- PMCID: PMC9794742
- DOI: 10.3389/fpls.2022.1011565
Genome-wide identification of Reverse Transcriptase domains of recently inserted endogenous plant pararetrovirus (Caulimoviridae)
Abstract
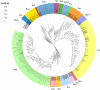
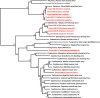
Endogenous viral elements (EVEs) are viral sequences that have been integrated into the nuclear chromosomes. Endogenous pararetrovirus (EPRV) are a class of EVEs derived from DNA viruses of the family Caulimoviridae. Previous works based on a limited number of genome assemblies demonstrated that EPRVs are abundant in plants and are present in several species. The availability of genome sequences has been immensely increased in the recent years and we took advantage of these resources to have a more extensive view of the presence of EPRVs in plant genomes. We analyzed 278 genome assemblies corresponding to 267 species (254 from Viridiplantae) using tBLASTn against a collection of conserved domains of the Reverse Transcriptases (RT) of Caulimoviridae. We concentrated our search on complete and well-conserved RT domains with an uninterrupted ORF comprising the genetic information for at least 300 amino acids. We obtained 11.527 sequences from the genomes of 202 species spanning the whole Tracheophyta clade. These elements were grouped in 57 clusters and classified in 13 genera, including a newly proposed genus we called Wendovirus. Wendoviruses are characterized by the presence of four open reading frames and two of them encode for aspartic proteinases. Comparing plant genomes, we observed important differences between the plant families and genera in the number and type of EPRVs found. In general, florendoviruses are the most abundant and widely distributed EPRVs. The presence of multiple identical RT domain sequences in some of the genomes suggests their recent amplification.
Keywords: Caulimoviridae; Reverse Transcriptase (RT); endogenous; pararetrovirus; virus.
Copyright © 2022 de Tomás and Vicient.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

