Interplay between rhizospheric Pseudomonas chlororaphis strains lays the basis for beneficial bacterial consortia
- PMID: 36589057
- PMCID: PMC9797978
- DOI: 10.3389/fpls.2022.1063182
Interplay between rhizospheric Pseudomonas chlororaphis strains lays the basis for beneficial bacterial consortia
Abstract
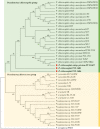
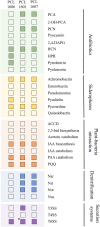
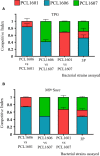
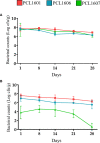
Pseudomonas chlororaphis (Pc) representatives are found as part of the rhizosphere-associated microbiome, and different rhizospheric Pc strains frequently perform beneficial activities for the plant. In this study we described the interactions between the rhizospheric Pc strains PCL1601, PCL1606 and PCL1607 with a focus on their effects on root performance. Differences among the three rhizospheric Pc strains selected were first observed in phylogenetic studies and confirmed by genome analysis, which showed variation in the presence of genes related to antifungal compounds or siderophore production, among others. Observation of the interactions among these strains under lab conditions revealed that PCL1606 has a better adaptation to environments rich in nutrients, and forms biofilms. Interaction experiments on plant roots confirmed the role of the different phenotypes in their lifestyle. The PCL1606 strain was the best adapted to the habitat of avocado roots, and PCL1607 was the least, and disappeared from the plant root scenario after a few days of interaction. These results confirm that 2 out 3 rhizospheric Pc strains were fully compatible (PCL1601 and PCL1606), efficiently colonizing avocado roots and showing biocontrol activity against the fungal pathogen Rosellinia necatrix. The third strain (PCL1607) has colonizing abilities when it is alone on the root but displayed difficulties under the competition scenario, and did not cause deleterious effects on the other Pc competitors when they were present. These results suggest that strains PCL1601 and PCL1606 are very well adapted to the avocado root environment and could constitute a basis for constructing a more complex beneficial microbial synthetic community associated with avocado plant roots.
Keywords: Pseudomonas chlororaphis; avocado; competitive index; consortia; rhizosphere; root colonization; syncom.
Copyright © 2022 Villar-Moreno, Tienda, Gutiérrez-Barranquero, Carrión, de Vicente, Cazorla and Arrebola.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

