Functions and evolution of FAM111 serine proteases
- PMID: 36589246
- PMCID: PMC9798293
- DOI: 10.3389/fmolb.2022.1081166
Functions and evolution of FAM111 serine proteases
Abstract
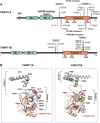
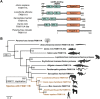
Proteolysis plays fundamental and regulatory roles in diverse cellular processes. The serine protease FAM111A (FAM111 trypsin-like peptidase A) emerged recently as a protease involved in two seemingly distinct processes: DNA replication and antiviral defense. FAM111A localizes to nascent DNA and plays a role at the DNA replication fork. At the fork, FAM111A is hypothesized to promote DNA replication at DNA-protein crosslinks (DPCs) and protein obstacles. On the other hand, FAM111A has also been identified as a host restriction factor for mutants of SV40 and orthopoxviruses. FAM111A also has a paralog, FAM111B, a serine protease with unknown cellular functions. Furthermore, heterozygous missense mutations in FAM111A and FAM111B cause distinct genetic disorders. In this review, we discuss possible models that could explain how FAM111A can function as a protease in both DNA replication and antiviral defense. We also review the consequences of FAM111A and FAM111B mutations and explore possible mechanisms underlying the diseases. Additionally, we propose a possible explanation for what drove the evolution of FAM111 proteins and discuss why some species have two FAM111 proteases. Altogether, studies of FAM111 proteases in DNA repair, antiviral defense, and genetic diseases will help us elucidate their functions and the regulatory mechanisms.
Keywords: DNA-protein crosslink (DPC); FAM111A; FAM111B; Kenny-Caffey Syndrome (KCS); POIKTMP; gracile bone dysplasia; protease; viral replication.
Copyright © 2022 Welter and Machida.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
E2F3-dependent activation of FAM111B restricts mouse cytomegalovirus replication in primate cells.J Virol. 2024 Dec 17;98(12):e0134924. doi: 10.1128/jvi.01349-24. Epub 2024 Nov 4. J Virol. 2024. PMID: 39494906 Free PMC article.
-
Unravelling the Intricate Roles of FAM111A and FAM111B: From Protease-Mediated Cellular Processes to Disease Implications.Int J Mol Sci. 2024 Feb 29;25(5):2845. doi: 10.3390/ijms25052845. Int J Mol Sci. 2024. PMID: 38474092 Free PMC article. Review.
-
FAM111 protease activity undermines cellular fitness and is amplified by gain-of-function mutations in human disease.EMBO Rep. 2020 Oct 5;21(10):e50662. doi: 10.15252/embr.202050662. Epub 2020 Aug 9. EMBO Rep. 2020. PMID: 32776417 Free PMC article.
-
Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.Nat Commun. 2024 Mar 7;15(1):2064. doi: 10.1038/s41467-024-46207-w. Nat Commun. 2024. PMID: 38453899 Free PMC article.
-
Case report: Late middle-aged features of FAM111A variant, Kenny-Caffey syndrome type 2-suggestive symptoms during a long follow-up.Front Endocrinol (Lausanne). 2023 Jan 4;13:1073173. doi: 10.3389/fendo.2022.1073173. eCollection 2022. Front Endocrinol (Lausanne). 2023. PMID: 36686468 Free PMC article. Review.
Cited by
-
Overexpressed FAM111B degrades GSDMA to promote esophageal cancer tumorigenesis and cisplatin resistance.Cell Oncol (Dordr). 2024 Feb;47(1):343-359. doi: 10.1007/s13402-023-00871-0. Epub 2023 Sep 6. Cell Oncol (Dordr). 2024. PMID: 37672204
-
Kenny-Caffey Syndrome Type 2 (KCS2): A New Case Report and Patient Follow-Up Optimization.J Clin Med. 2024 Dec 28;14(1):118. doi: 10.3390/jcm14010118. J Clin Med. 2024. PMID: 39797201 Free PMC article.
-
N6 -methyladenosine-modified FAM111A-DT promotes hepatocellular carcinoma growth via epigenetically activating FAM111A.Cancer Sci. 2023 Sep;114(9):3649-3665. doi: 10.1111/cas.15886. Epub 2023 Jul 3. Cancer Sci. 2023. PMID: 37400994 Free PMC article.
-
E2F3-dependent activation of FAM111B restricts mouse cytomegalovirus replication in primate cells.J Virol. 2024 Dec 17;98(12):e0134924. doi: 10.1128/jvi.01349-24. Epub 2024 Nov 4. J Virol. 2024. PMID: 39494906 Free PMC article.
-
Editorial: The repair of DNA-protein crosslinks.Front Mol Biosci. 2023 Apr 28;10:1203479. doi: 10.3389/fmolb.2023.1203479. eCollection 2023. Front Mol Biosci. 2023. PMID: 37187895 Free PMC article. No abstract available.
References
-
- Alabert C., Bukowski-Wills J. C., Lee S. B., Kustatscher G., Nakamura K., de Lima Alves F., et al. (2014). Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16 (3), 281–293. 10.1038/ncb2918 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

