Biological properties of the BCL-2 family protein BCL-RAMBO, which regulates apoptosis, mitochondrial fragmentation, and mitophagy
- PMID: 36589739
- PMCID: PMC9800997
- DOI: 10.3389/fcell.2022.1065702
Biological properties of the BCL-2 family protein BCL-RAMBO, which regulates apoptosis, mitochondrial fragmentation, and mitophagy
Abstract
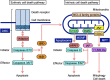
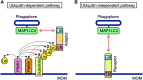
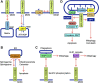
Mitochondria play an essential role in the regulation of cellular stress responses, including cell death. Damaged mitochondria are removed by fission and fusion cycles and mitophagy, which counteract cell death. BCL-2 family proteins possess one to four BCL-2 homology domains and regulate apoptosis signaling at mitochondria. BCL-RAMBO, also known as BCL2-like 13 (BCL2L13), was initially identified as one of the BCL-2 family proteins inducing apoptosis. Mitophagy receptors recruit the ATG8 family proteins MAP1LC3/GABARAP via the MAP1LC3-interacting region (LIR) motif to initiate mitophagy. In addition to apoptosis, BCL-RAMBO has recently been identified as a mitophagy receptor that possesses the LIR motif and regulates mitochondrial fragmentation and mitophagy. In the 20 years since its discovery, many important findings on BCL-RAMBO have been increasingly reported. The biological properties of BCL-RAMBO are reviewed herein.
Keywords: BCL-RAMBO; BCL2L13; apoptosis; cell death; miRNAs; mitochondrial fragmentation; mitophagy; phosphorylation.
Copyright © 2022 Kataoka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
PGAM5 interacts with Bcl-rambo and regulates apoptosis and mitophagy.Exp Cell Res. 2022 Nov 1;420(1):113342. doi: 10.1016/j.yexcr.2022.113342. Epub 2022 Sep 6. Exp Cell Res. 2022. PMID: 36075447
-
Selective binding of mitophagy receptor protein Bcl-rambo to LC3/GABARAP family proteins.Biochem Biophys Res Commun. 2020 Sep 10;530(1):292-300. doi: 10.1016/j.bbrc.2020.07.039. Epub 2020 Aug 6. Biochem Biophys Res Commun. 2020. PMID: 32828302
-
The human Bcl-2 family member Bcl-rambo and voltage-dependent anion channels manifest a genetic interaction in Drosophila and cooperatively promote the activation of effector caspases in human cultured cells.Exp Cell Res. 2019 Aug 15;381(2):223-234. doi: 10.1016/j.yexcr.2019.05.015. Epub 2019 May 15. Exp Cell Res. 2019. PMID: 31102594
-
BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32.Autophagy. 2015;11(10):1932-3. doi: 10.1080/15548627.2015.1084459. Autophagy. 2015. PMID: 26506896 Free PMC article. Review.
-
BCL2L13: physiological and pathological meanings.Cell Mol Life Sci. 2021 Mar;78(6):2419-2428. doi: 10.1007/s00018-020-03702-9. Epub 2020 Nov 17. Cell Mol Life Sci. 2021. PMID: 33201252 Free PMC article. Review.
Cited by
-
1-L Transcription of SARS-CoV-2 Spike Protein S1 Subunit.Int J Mol Sci. 2024 Apr 18;25(8):4440. doi: 10.3390/ijms25084440. Int J Mol Sci. 2024. PMID: 38674024 Free PMC article.
-
The BCL-2 family protein BCL-RAMBO interacts and cooperates with GRP75 to promote its apoptosis signaling pathway.Sci Rep. 2023 Aug 28;13(1):14041. doi: 10.1038/s41598-023-41196-0. Sci Rep. 2023. PMID: 37640805 Free PMC article.
-
The role of Bcl‑2 in controlling the transition between autophagy and apoptosis (Review).Mol Med Rep. 2025 Jul;32(1):172. doi: 10.3892/mmr.2025.13537. Epub 2025 Apr 17. Mol Med Rep. 2025. PMID: 40242969 Free PMC article. Review.
-
MG53 deficiency mediated skeletal muscle dysfunction in chronic obstructive pulmonary disease via impairing mitochondrial fission.Redox Biol. 2025 Jun;83:103663. doi: 10.1016/j.redox.2025.103663. Epub 2025 May 3. Redox Biol. 2025. PMID: 40345073 Free PMC article.
-
BCL2L13 at endoplasmic reticulum-mitochondria contact sites regulates calcium homeostasis to maintain skeletal muscle function.iScience. 2024 Jul 14;27(8):110510. doi: 10.1016/j.isci.2024.110510. eCollection 2024 Aug 16. iScience. 2024. PMID: 39175772 Free PMC article.
References
-
- Aryal S., Anand D., Hernandez F. G., Weaterbee B. A., Huang H., Reddy A. P., et al. (2020). MS/MS in silico subtraction-based proteomic profiling as an approach to facilitate disease gene discovery: application to lens development and cataract. Hum. Genet. 139, 151–184. 10.1007/s00439-019-02095-5 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

