Construction of a prognostic assessment model for colon cancer patients based on immune-related genes and exploration of related immune characteristics
- PMID: 36589748
- PMCID: PMC9800979
- DOI: 10.3389/fcell.2022.993580
Construction of a prognostic assessment model for colon cancer patients based on immune-related genes and exploration of related immune characteristics
Abstract
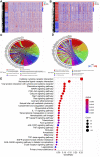
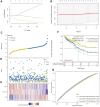
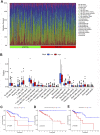
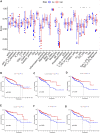
Objectives: To establish a novel risk score model that could predict the survival and immune response of patients with colon cancer. Methods: We used The Cancer Genome Atlas (TCGA) database to get mRNA expression profile data, corresponding clinical information and somatic mutation data of patients with colon cancer. Limma R software package and univariate Cox regression were performed to screen out immune-related prognostic genes. GO (Gene ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) were used for gene function enrichment analysis. The risk scoring model was established by Lasso regression and multivariate Cox regression. CIBERSORT was conducted to estimate 22 types of tumor-infiltrating immune cells and immune cell functions in tumors. Correlation analysis was used to demonstrate the relationship between the risk score and immune escape potential. Results: 679 immune-related genes were selected from 7846 differentially expressed genes (DEGs). GO and KEGG analysis found that immune-related DEGs were mainly enriched in immune response, complement activation, cytokine-cytokine receptor interaction and so on. Finally, we established a 3 immune-related genes risk scoring model, which was the accurate independent predictor of overall survival (OS) in colon cancer. Correlation analysis indicated that there were significant differences in T cell exclusion potential in low-risk and high-risk groups. Conclusion: The immune-related gene risk scoring model could contribute to predicting the clinical outcome of patients with colon cancer.
Keywords: colon cancer; immune cell infiltration; immune escape; immune-related gene; risk model.
Copyright © 2022 Wan, He, Yang, Cheng, Li, Zhang, Zhang, Dai, Yu, Li, Xiong and Wan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

