Detection of ROS1 gene fusions using next-generation sequencing for patients with malignancy in China
- PMID: 36589752
- PMCID: PMC9798300
- DOI: 10.3389/fcell.2022.1035033
Detection of ROS1 gene fusions using next-generation sequencing for patients with malignancy in China
Abstract
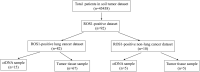
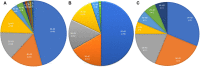
Objective: This study aimed to identify ROS1 fusion partners in Chinese patients with solid tumors. Methods: Next-generation sequencing (NGS) analysis was used to detect ROS1 rearrangement in 45,438 Chinese patients with solid tumors between 2015 and 2020, and the clinical characteristics and genetic features of gene fusion were evaluated. H&E staining of the excised tumor tissues was conducted. Samples with a tumor cell content ≥ 20% were included for subsequent DNA extraction and sequencing analysis. Results: A total of 92 patients with ROS1 rearrangements were identified using next-generation sequencing, and the most common histological type lung cancer. From the 92 ROS1 fusion cases, 24 ROS1 fusion partners had been identified, including 14 novel partners and 10 reported partners. Of these, CD74, EZR, SDC4, and TPM3 were the four most frequently occurring partners. Fourteen novel ROS1 fusion partners were detected in 16 patients, including DCBLD1-ROS1, FRK-ROS1, and VGLL2-ROS1. In many patients, the ROS1 breakpoint was located between exons 32 and 34. Conclusion: This study describes 14 novel ROS1 fusion partners based on the largest ROS1 fusion cohort, and the ROS1 breakpoint was mostly located between exons 32 and 34. Additionally, next-generation sequencing is an optional method for identifying novel ROS1 fusions.
Keywords: ROS1 breakpoint; ROS1 fusions partners; lung cancer; next-generation sequencing; solid tumor.
Copyright © 2022 Li, Chen, Huang, Zhang, Hu, Jiao and Quan.
Conflict of interest statement
Authors DZ and MH were employed by the company 3D Medicines Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Anjum R., Zhang S., Squillace R., Clackson T., Garner A. P., Rivera V. M. (2012). 164 the dual ALK/EGFR inhibitor AP26113 also potently inhibits. Eur. J. Cancer 48, 50. 10.1016/s0959-8049(12)71962-7 - DOI
-
- Birch A. H., Arcand S. L., Oros K. K., Rahimi K., Watters A. K., Provencher D., et al. (2011). Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PloS one 6 (12), e28250. 10.1371/journal.pone.0028250 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

