A DNA biosensors-based microfluidic platform for attomolar real-time detection of unamplified SARS-CoV-2 virus
- PMID: 36589921
- PMCID: PMC9793959
- DOI: 10.1016/j.biosx.2022.100302
A DNA biosensors-based microfluidic platform for attomolar real-time detection of unamplified SARS-CoV-2 virus
Abstract
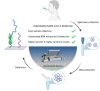
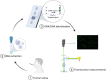
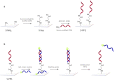
The emergence of the coronavirus 2019 (COVID-19) arose the need for rapid, accurate and massive virus detection methods to control the spread of infectious diseases. In this work, a device, deployable in non-medical environments, has been developed for the detection of non-amplified SARS-CoV-2 RNA. A SARS-CoV-2 specific probe was designed and covalently immobilized at the surface of glass slides to fabricate a DNA biosensor. The resulting system was integrated in a microfluidic platform, in which viral RNA was extracted from non-treated human saliva, before hybridizing at the surface of the sensor. The formed DNA/RNA duplex was detected in presence of SYBR Green I using an opto-electronic system, based on a high-power LED and a photo multiplier tube, which convert the emitted fluorescence into an electrical signal that can be processed in less than 10 min. The limit of detection of the resulting microfluidic platform reached six copies of viral RNA per microliter of sample (equal to 10 aM) and satisfied the safety margin. The absence of non-specific adsorption and the selectivity for SARS-CoV-2 RNA were established. In addition, the designed device could be applicable for the detection of a variety of viruses by simple modification of the immobilized probe.
Keywords: DNA-biosensor; Fluorescence; Microfluidic; RNA extraction; SARS-CoV-2 detection; Silica slide.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Bidar N., Amini M., Oroojalian F., Baradaran B., Hosseini S.S., Shahbazi M.-A., Hashemzaei M., Mokhtarzadeh A., Hamblin M.R., de la Guardia M. Trends Anal. Chem. 2021;134
-
- Brown L.J., Cummins J., Hamilton A., Brown T. Chem. Commun. 2000;7:621–622.
-
- Bruch R., Johnston M., Kling A., Mattmüller T., Baaske J., Partel S., Madlener S., Weber W., Urban G.A., Dincer C. Biosens. Bioelectron. 2021;177 - PubMed
-
- Bryant J. Analog. Dialogue. 2014;48:1–3.
LinkOut - more resources
Full Text Sources
Miscellaneous
