Elucidating genes and gene networks linked to individual susceptibility to milk fat depression in dairy goats
- PMID: 36590804
- PMCID: PMC9798324
- DOI: 10.3389/fvets.2022.1037764
Elucidating genes and gene networks linked to individual susceptibility to milk fat depression in dairy goats
Abstract
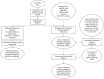
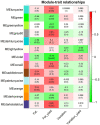
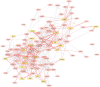
Dietary supplementation with marine lipids modulates ruminant milk composition toward a healthier fatty acid profile for consumers, but it also causes milk fat depression (MFD). Because the dairy goat industry is mainly oriented toward cheese manufacturing, MFD can elicit economic losses. There is large individual variation in animal susceptibility with goats more (RESPO+) or less (RESPO-) responsive to diet-induced MFD. Thus, we used RNA-Seq to examine gene expression profiles in mammary cells to elucidate mechanisms underlying MFD in goats and individual variation in the extent of diet-induced MFD. Differentially expression analyses (DEA) and weighted gene co-expression network analysis (WGCNA) of RNA-Seq data were used to study milk somatic cell transcriptome changes in goats consuming a diet supplemented with marine lipids. There were 45 differentially expressed genes (DEGs) between control (no-MFD, before diet-induced MFD) and MFD, and 18 between RESPO+ and RESPO-. Biological processes and pathways such as "RNA transcription" and "Chromatin modifying enzymes" were downregulated in MFD compared with controls. Regarding susceptibility to diet-induced MFD, we identified the "Triglyceride Biosynthesis" pathway upregulated in RESPO- goats. The WGCNA approach identified 9 significant functional modules related to milk fat production and one module to the fat yield decrease in diet-induced MFD. The onset of MFD in dairy goats is influenced by the downregulation of SREBF1, other transcription factors and chromatin-modifying enzymes. A list of DEGs between RESPO+ and RESPO- goats (e.g., DBI and GPD1), and a co-related gene network linked to the decrease in milk fat (ABCD3, FABP3, and PLIN2) was uncovered. Results suggest that alterations in fatty acid transport may play an important role in determining individual variation. These candidate genes should be further investigated.
Keywords: RNA-Seq; WGCNA; goat; individual susceptibility; milk-fat depression; nutrigenomics.
Copyright © 2022 Suárez-Vega, Gutiérrez-Gil, Toral, Frutos, Loor, Arranz and Hervás.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bauman DE, Tyburczy C, O'Donnell AM, Lock AL. Milklipids: Conjugated linoleic acid. In:Fuquay JM, Fox PF, McSweeney PLH, editors. Encyclopedia of Dairy Sciences. 2nd ed. San Diego, CA: Academic Press; (2011). p. 660–4.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

