Network analysis uncovers the communication structure of SARS-CoV-2 spike protein identifying sites for immunogen design
- PMID: 36590900
- PMCID: PMC9791713
- DOI: 10.1016/j.isci.2022.105855
Network analysis uncovers the communication structure of SARS-CoV-2 spike protein identifying sites for immunogen design
Abstract
The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has triggered myriad efforts to understand the structure and dynamics of this complex pathogen. The spike glycoprotein of SARS-CoV-2 is a significant target for immunogens as it is the means by which the virus enters human cells, while simultaneously sporting mutations responsible for immune escape. These functional and escape processes are regulated by complex molecular-level interactions. Our study presents quantitative insights on domain and residue contributions to allosteric communication, immune evasion, and local- and global-level control of functions through the derivation of a weighted graph representation from all-atom MD simulations. Focusing on the ancestral form and the D614G-variant, we provide evidence of the utility of our approach by guiding the selection of a mutation that alters the spike's stability. Taken together, the network approach serves as a valuable tool to evaluate communication "hot-spots" in proteins to guide design of stable immunogens.
Keywords: Biological sciences; Immunology; Structural biology; Virology.
© 2022.
Conflict of interest statement
The authors declare no competing interests.
Figures
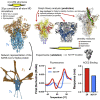

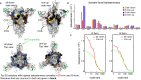


Similar articles
-
Integrating Conformational Dynamics and Perturbation-Based Network Modeling for Mutational Profiling of Binding and Allostery in the SARS-CoV-2 Spike Variant Complexes with Antibodies: Balancing Local and Global Determinants of Mutational Escape Mechanisms.Biomolecules. 2022 Jul 10;12(7):964. doi: 10.3390/biom12070964. Biomolecules. 2022. PMID: 35883520 Free PMC article.
-
Computational analysis of protein stability and allosteric interaction networks in distinct conformational forms of the SARS-CoV-2 spike D614G mutant: reconciling functional mechanisms through allosteric model of spike regulation.J Biomol Struct Dyn. 2022;40(20):9724-9741. doi: 10.1080/07391102.2021.1933594. Epub 2021 Jun 1. J Biomol Struct Dyn. 2022. PMID: 34060425
-
Computer Simulations and Network-Based Profiling of Binding and Allosteric Interactions of SARS-CoV-2 Spike Variant Complexes and the Host Receptor: Dissecting the Mechanistic Effects of the Delta and Omicron Mutations.Int J Mol Sci. 2022 Apr 15;23(8):4376. doi: 10.3390/ijms23084376. Int J Mol Sci. 2022. PMID: 35457196 Free PMC article.
-
D614G mutation and SARS-CoV-2: impact on S-protein structure, function, infectivity, and immunity.Appl Microbiol Biotechnol. 2021 Dec;105(24):9035-9045. doi: 10.1007/s00253-021-11676-2. Epub 2021 Nov 10. Appl Microbiol Biotechnol. 2021. PMID: 34755213 Free PMC article. Review.
-
Structural Plasticity and Immune Evasion of SARS-CoV-2 Spike Variants.Viruses. 2022 Jun 9;14(6):1255. doi: 10.3390/v14061255. Viruses. 2022. PMID: 35746726 Free PMC article. Review.
Cited by
-
Complementary Pocket and Network-Based Approach to Search for Spike Protein Allosteric Pocket Sites.ACS Omega. 2023 Oct 3;8(48):45313-45325. doi: 10.1021/acsomega.3c04007. eCollection 2023 Dec 5. ACS Omega. 2023. PMID: 38075758 Free PMC article.
-
Evolution of the SARS-CoV-2 Omicron spike.Cell Rep. 2023 Dec 26;42(12):113444. doi: 10.1016/j.celrep.2023.113444. Epub 2023 Nov 18. Cell Rep. 2023. PMID: 37979169 Free PMC article. Review.
-
Paired associated SARS-CoV-2 spike variable positions: a network analysis approach to emerging variants.mSystems. 2023 Aug 31;8(4):e0044023. doi: 10.1128/msystems.00440-23. Epub 2023 Jul 11. mSystems. 2023. PMID: 37432011 Free PMC article.
-
Evolutionary rewiring of the dynamic network underpinning allosteric epistasis in NS1 of the influenza A virus.Proc Natl Acad Sci U S A. 2025 Feb 25;122(8):e2410813122. doi: 10.1073/pnas.2410813122. Epub 2025 Feb 20. Proc Natl Acad Sci U S A. 2025. PMID: 39977319
-
Predicting permeation of compounds across the outer membrane of P. aeruginosa using molecular descriptors.Commun Chem. 2024 Apr 12;7(1):84. doi: 10.1038/s42004-024-01161-y. Commun Chem. 2024. PMID: 38609430 Free PMC article.
References
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. https://europepmc.org/articles/PMC7095418 - DOI - PMC - PubMed
-
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265—269. doi: 10.1038/s41586-020-2008-3. https://europepmc.org/articles/PMC7094943 - DOI - PMC - PubMed
-
- Who coronavirus (covid-19) dashboard (latest seen on 11/23/2022 at 12:23 pm et) 2022. https://covid19.who.int/
-
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. https://europepmc.org/articles/PMC4860016 - DOI - PMC - PubMed
-
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the sars-cov-2 spike glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. https://europepmc.org/articles/PMC7833104 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

