Recent changes in the mutational dynamics of the SARS-CoV-2 main protease substantiate the danger of emerging resistance to antiviral drugs
- PMID: 36590977
- PMCID: PMC9794616
- DOI: 10.3389/fmed.2022.1061142
Recent changes in the mutational dynamics of the SARS-CoV-2 main protease substantiate the danger of emerging resistance to antiviral drugs
Abstract
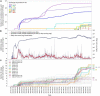
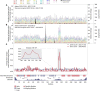
Introduction: The current coronavirus pandemic is being combated worldwide by nontherapeutic measures and massive vaccination programs. Nevertheless, therapeutic options such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main-protease (Mpro) inhibitors are essential due to the ongoing evolution toward escape from natural or induced immunity. While antiviral strategies are vulnerable to the effects of viral mutation, the relatively conserved Mpro makes an attractive drug target: Nirmatrelvir, an antiviral targeting its active site, has been authorized for conditional or emergency use in several countries since December 2021, and a number of other inhibitors are under clinical evaluation. We analyzed recent SARS-CoV-2 genomic data, since early detection of potential resistances supports a timely counteraction in drug development and deployment, and discovered accelerated mutational dynamics of Mpro since early December 2021.
Methods: We performed a comparative analysis of 10.5 million SARS-CoV-2 genome sequences available by June 2022 at GISAID to the NCBI reference genome sequence NC_045512.2. Amino-acid exchanges within high-quality regions in 69,878 unique Mpro sequences were identified and time- and in-depth sequence analyses including a structural representation of mutational dynamics were performed using in-house software.
Results: The analysis showed a significant recent event of mutational dynamics in Mpro. We report a remarkable increase in mutational variability in an eight-residue long consecutive region (R188-G195) near the active site since December 2021.
Discussion: The increased mutational variability in close proximity to an antiviral-drug binding site as described herein may suggest the onset of the development of antiviral resistance. This emerging diversity urgently needs to be further monitored and considered in ongoing drug development and lead optimization.
Keywords: COVID-19; Mpro; Paxlovid; SARS-CoV-2; drug resistance; main protease; nirmatrelvir; viral evolution.
Copyright © 2022 Parigger, Krassnigg, Schopper, Singh, Tappler, Köchl, Hetmann, Gruber, Steinkellner and Gruber.
Conflict of interest statement
LP, AK, TS, MH, KK, and AS report working for Innophore. KG, GS, and CG report being shareholders of Innophore GmbH, an enzyme and drug discovery company. Additionally, GS and CG report being managing directors of Innophore. The research described here is scientifically and financially independent of the efforts in the above-mentioned company Innophore and open science. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Deng X, StJohn S, Osswald H, O’Brien A, Banach B, Sleeman K, et al. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J Virol. (2014) 88:11886–98. 10.1128/JVI.01528-14 - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

