Host antiviral factors hijack furin to block SARS-CoV-2, ebola virus, and HIV-1 glycoproteins cleavage
- PMID: 36591809
- PMCID: PMC9897805
- DOI: 10.1080/22221751.2022.2164742
Host antiviral factors hijack furin to block SARS-CoV-2, ebola virus, and HIV-1 glycoproteins cleavage
Abstract
Viral envelope glycoproteins are crucial for viral infections. In the process of enveloped viruses budding and release from the producer cells, viral envelope glycoproteins are presented on the viral membrane surface as spikes, promoting the virus's next-round infection of target cells. However, the host cells evolve counteracting mechanisms in the long-term virus-host co-evolutionary processes. For instance, the host cell antiviral factors could potently suppress viral replication by targeting their envelope glycoproteins through multiple channels, including their intracellular synthesis, glycosylation modification, assembly into virions, and binding to target cell receptors. Recently, a group of studies discovered that some host antiviral proteins specifically recognized host proprotein convertase (PC) furin and blocked its cleavage of viral envelope glycoproteins, thus impairing viral infectivity. Here, in this review, we briefly summarize several such host antiviral factors and analyze their roles in reducing furin cleavage of viral envelope glycoproteins, aiming at providing insights for future antiviral studies.
Keywords: Antiviral factors; SARS-CoV-2; cleavage; furin; viral glycoprotein.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
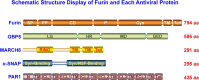
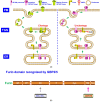

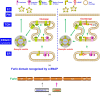
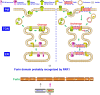
References
-
- Poranen MM, Daugelavicius R, Bamford DH.. Common principles in viral entry. Annu Rev Microbiol. 2002;56:521–538. - PubMed
-
- Lu L, Su S, Yang H, et al. . Antivirals with common targets against highly pathogenic viruses. Cell. 2021 Mar 18;184(6):1604–1620. - PubMed
-
- Wei W, Yu XF.. HIV-1 envelope under attack. Trends Microbiol. 2016 Mar;24(3):164–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
