A liquid-to-solid phase transition of Cu/Zn superoxide dismutase 1 initiated by oxidation and disease mutation
- PMID: 36592929
- PMCID: PMC9898760
- DOI: 10.1016/j.jbc.2022.102857
A liquid-to-solid phase transition of Cu/Zn superoxide dismutase 1 initiated by oxidation and disease mutation
Abstract
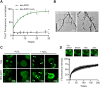
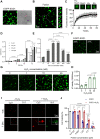
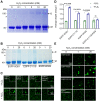
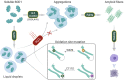
Cu/Zn superoxide dismutase 1 (SOD1) has a high propensity to misfold and form abnormal aggregates when it is subjected to oxidative stress or carries mutations associated with amyotrophic lateral sclerosis. However, the transition from functional soluble SOD1 protein to aggregated SOD1 protein is not completely clear. Here, we propose that liquid-liquid phase separation (LLPS) represents a biophysical process that converts soluble SOD1 into aggregated SOD1. We determined that SOD1 undergoes LLPS in vitro and cells under oxidative stress. Abnormal oxidation of SOD1 induces maturation of droplets formed by LLPS, eventually leading to protein aggregation and fibrosis, and involves residues Cys111 and Trp32. Additionally, we found that pathological mutations in SOD1 associated with ALS alter the morphology and material state of the droplets and promote the transformation of SOD1 to solid-like oligomers which are toxic to nerve cells. Furthermore, the fibrous aggregates formed by both pathways have a concentration-dependent toxicity effect on nerve cells. Thus, these combined results strongly indicate that LLPS may play a major role in pathological SOD1 aggregation, contributing to pathogenesis in ALS.
Keywords: SOD1; aggregation; liquid-liquid phase separation; neurodegenerative diseases; oxidation.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Bruijn L.I., Houseweart M.K., Kato S., Anderson K.L., Anderson S.D., Ohama E., et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. - PubMed
-
- Bruijn L.I., Becher M.W., Lee M.K., Anderson K.L., Jenkins N.A., Copeland N.G., et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. - PubMed
-
- Cleveland D.W., Rothstein J.D. From charcot to lou gehrig: deciphering selective motor neuron death in als. Nat. Rev. Neurosci. 2001;2:806–819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

