MicroRNA-223 attenuates hepatocarcinogenesis by blocking hypoxia-driven angiogenesis and immunosuppression
- PMID: 36593103
- PMCID: PMC11283862
- DOI: 10.1136/gutjnl-2022-327924
MicroRNA-223 attenuates hepatocarcinogenesis by blocking hypoxia-driven angiogenesis and immunosuppression
Abstract
Objective: The current treatment for hepatocellular carcinoma (HCC) to block angiogenesis and immunosuppression provides some benefits only for a subset of patients with HCC, thus optimised therapeutic regimens are unmet needs, which require a thorough understanding of the underlying mechanisms by which tumour cells orchestrate an inflamed tumour microenvironment with significant myeloid cell infiltration. MicroRNA-223 (miR-223) is highly expressed in myeloid cells but its role in regulating tumour microenvironment remains unknown.
Design: Wild-type and miR-223 knockout mice were subjected to two mouse models of inflammation-associated HCC induced by injection of diethylnitrosamine (DEN) or orthotopic HCC cell implantation in chronic carbon tetrachloride (CCl4)-treated mice.
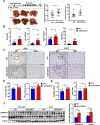
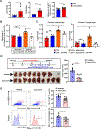
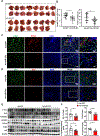
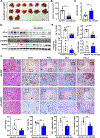
Results: Genetic deletion of miR-223 markedly exacerbated tumourigenesis in inflammation-associated HCC. Compared with wild-type mice, miR-223 knockout mice had more infiltrated programmed cell death 1 (PD-1+) T cells and programmed cell death ligand 1 (PD-L1+) macrophages after DEN+CCl4 administration. Bioinformatic analyses of RNA sequencing data revealed a strong correlation between miR-223 levels and tumour hypoxia, a condition that is well-documented to regulate PD-1/PD-L1. In vivo and in vitro mechanistic studies demonstrated that miR-223 did not directly target PD-1 and PD-L1 in immune cells rather than indirectly downregulated them by modulating tumour microenvironment via the suppression of hypoxia-inducible factor 1α-driven CD39/CD73-adenosine pathway in HCC. Moreover, gene delivery of miR-223 via adenovirus inhibited angiogenesis and hypoxia-mediated PD-1/PD-L1 activation in both HCC models, thereby hindering HCC progression.
Conclusion: The miR-223 plays a critical role in modulating hypoxia-induced tumour immunosuppression and angiogenesis, which may serve as a novel therapeutic target for HCC.
Keywords: CANCER IMMUNOBIOLOGY; HEPATOCELLULAR CARCINOMA; INFLAMMATORY CELLS.
© Author(s) (or their employer(s)) 2023. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures







Comment in
-
MicroRNA-223: a key regulator of liver tumour microenvironment.Gut. 2023 Oct;72(10):1811-1812. doi: 10.1136/gutjnl-2022-329322. Epub 2023 Feb 15. Gut. 2023. PMID: 36792357 No abstract available.
Similar articles
-
E-twenty-six-specific sequence variant 5 (ETV5) facilitates hepatocellular carcinoma progression and metastasis through enhancing polymorphonuclear myeloid-derived suppressor cell (PMN-MDSC)-mediated immunosuppression.Gut. 2025 Jun 6;74(7):1137-1149. doi: 10.1136/gutjnl-2024-333944. Gut. 2025. PMID: 40015948
-
Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade.Gut. 2019 Sep;68(9):1653-1666. doi: 10.1136/gutjnl-2019-318419. Epub 2019 Mar 22. Gut. 2019. PMID: 30902885
-
Blocking Triggering Receptor Expressed on Myeloid Cells-1-Positive Tumor-Associated Macrophages Induced by Hypoxia Reverses Immunosuppression and Anti-Programmed Cell Death Ligand 1 Resistance in Liver Cancer.Hepatology. 2019 Jul;70(1):198-214. doi: 10.1002/hep.30593. Epub 2019 Apr 12. Hepatology. 2019. PMID: 30810243 Free PMC article.
-
Viral status, immune microenvironment and immunological response to checkpoint inhibitors in hepatocellular carcinoma.J Immunother Cancer. 2020 Apr;8(1):e000394. doi: 10.1136/jitc-2019-000394. J Immunother Cancer. 2020. PMID: 32303615 Free PMC article.
-
Programmed cell death protein-1 (PD-1)/programmed death-ligand-1 (PD-L1) axis in hepatocellular carcinoma: prognostic and therapeutic perspectives.Clin Transl Oncol. 2019 Jun;21(6):702-712. doi: 10.1007/s12094-018-1975-4. Epub 2018 Nov 1. Clin Transl Oncol. 2019. PMID: 30387047 Review.
Cited by
-
Mechanism research of non-coding RNA in immune checkpoint inhibitors therapy.Cancer Sci. 2024 Nov;115(11):3520-3531. doi: 10.1111/cas.16309. Epub 2024 Aug 13. Cancer Sci. 2024. PMID: 39136293 Free PMC article. Review.
-
Co-delivery of camptothecin and MiR-145 by lipid nanoparticles for MRI-visible targeted therapy of hepatocellular carcinoma.J Exp Clin Cancer Res. 2024 Aug 30;43(1):247. doi: 10.1186/s13046-024-03167-9. J Exp Clin Cancer Res. 2024. PMID: 39215325 Free PMC article.
-
Hypoxia-induced tRF-3Thr-CGT promotes hepatocellular carcinoma progression via mitochondrial energy metabolism remodeling dependent on the mtDNA-translation mechanism.Front Pharmacol. 2025 May 30;16:1549373. doi: 10.3389/fphar.2025.1549373. eCollection 2025. Front Pharmacol. 2025. PMID: 40520188 Free PMC article.
-
Hypoxia: syndicating triple negative breast cancer against various therapeutic regimens.Front Oncol. 2023 Jul 10;13:1199105. doi: 10.3389/fonc.2023.1199105. eCollection 2023. Front Oncol. 2023. PMID: 37492478 Free PMC article. Review.
-
Lenvatinib Induces Immunogenic Cell Death and Triggers Toll-Like Receptor-3/4 Ligands in Hepatocellular Carcinoma.J Hepatocell Carcinoma. 2023 Apr 24;10:697-712. doi: 10.2147/JHC.S401639. eCollection 2023. J Hepatocell Carcinoma. 2023. PMID: 37138764 Free PMC article.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. - PubMed
-
- Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med 2020;382:1894–1905. - PubMed
-
- Sengez B, Carr BI, Alotaibi H. EMT and Inflammation: Crossroads in HCC. J Gastrointest Cancer 2022. - PubMed
-
- Geh D, Leslie J, Rumney R, et al. Neutrophils as potential therapeutic targets in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2022;19:257–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
