Magnify is a universal molecular anchoring strategy for expansion microscopy
- PMID: 36593399
- PMCID: PMC10264239
- DOI: 10.1038/s41587-022-01546-1
Magnify is a universal molecular anchoring strategy for expansion microscopy
Abstract
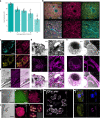
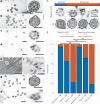
Expansion microscopy enables nanoimaging with conventional microscopes by physically and isotropically magnifying preserved biological specimens embedded in a crosslinked water-swellable hydrogel. Current expansion microscopy protocols require prior treatment with reactive anchoring chemicals to link specific labels and biomolecule classes to the gel. We describe a strategy called Magnify, which uses a mechanically sturdy gel that retains nucleic acids, proteins and lipids without the need for a separate anchoring step. Magnify expands biological specimens up to 11 times and facilitates imaging of cells and tissues with effectively around 25-nm resolution using a diffraction-limited objective lens of about 280 nm on conventional optical microscopes or with around 15 nm effective resolution if combined with super-resolution optical fluctuation imaging. We demonstrate Magnify on a broad range of biological specimens, providing insight into nanoscopic subcellular structures, including synaptic proteins from mouse brain, podocyte foot processes in formalin-fixed paraffin-embedded human kidney and defects in cilia and basal bodies in drug-treated human lung organoids.
© 2023. The Author(s).
Conflict of interest statement
The authors declare the following competing financial interest(s): Y.Z. and A.K. are inventors on several inventions related to ExM. The remaining authors declare no competing interests.
Figures




Comment in
-
Next-generation expansion microscopy.Nat Methods. 2023 Feb;20(2):175. doi: 10.1038/s41592-023-01793-3. Nat Methods. 2023. PMID: 36765137 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

