A proteomic profile of the healthy human placenta
- PMID: 36593452
- PMCID: PMC9808999
- DOI: 10.1186/s12014-022-09388-4
A proteomic profile of the healthy human placenta
Abstract
Background: The placenta remains one of the least studied organs within the human body. Yet, placental dysfunction has been associated with various pregnancy complications leading to both maternal and fetal death and long-term health consequences. The aim of this study was to characterise the protein networks of healthy term placental sub-anatomical regions using label free quantification mass spectrometry.
Methods: Three healthy placentae were sampled at five sample sites and each biopsy was dissected into maternal-, middle-, and fetal- sub-anatomical regions. Quadrupole-orbitrap mass spectrometer was used in data dependant analysis mode to identify 1859 unique proteins before detailed differential expression between regions.
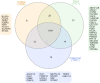
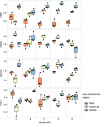
Results: Protein profiling identified 1081, 1086, and 1101 proteins in maternal, middle, and fetal sub-anatomical regions respectively. Differentially expressed proteins were identified considering the effect between sample site location and sub-anatomical region on protein expression. Of these, 374 differentially expressed proteins (Two-way ANOVA adjusted p-value < 0.05, HSD Tukey adjusted p-value 0.05) were identified between sample site locations and sub-anatomical regions. The placenta specific disease map NaviCenta ( https://www.sbi.uni-rostock.de/minerva/index.xhtml?id=NaviCenta ) was used to focus functional analysis results to the placenta specific context. Subsequently, functional analysis with a focus on senescence, and mitochondrial function were performed. Significant differences were observed between sub-anatomical regions in protein intensity and composition. A decrease in anti-senescent proteins within the maternal sub-anatomical region, and an increase in proteins associated with a switch from ATP to fatty acid consumption as a source of energy between middle and fetal sub-anatomical regions were observed.
Conclusion: These results suggest that normal proteomic variations exist within the anatomical structure of the placenta, thus recommending serial sectioning methodology for consistent placental research.
Keywords: Bioinformatics; Disease map; Network analysis; Placenta; Proteomics; Systems biology.
© 2023. The Author(s).
Conflict of interest statement
Authors report no conflict of Interest.
Figures



References
-
- Maltepe E, Penn AA. Development, Function, and Pathology of the Placenta. In: Gleason CA, Juul SE, editors. Avery's Diseases of the Newborn. 10. Philadelphia: Elsevier; 2018. pp. 40–60.e8.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
