Macrophages and microglia in glioblastoma: heterogeneity, plasticity, and therapy
- PMID: 36594466
- PMCID: PMC9797335
- DOI: 10.1172/JCI163446
Macrophages and microglia in glioblastoma: heterogeneity, plasticity, and therapy
Abstract
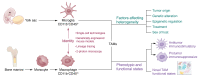
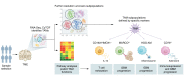
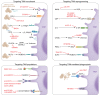
Glioblastoma (GBM) is the most aggressive tumor in the central nervous system and contains a highly immunosuppressive tumor microenvironment (TME). Tumor-associated macrophages and microglia (TAMs) are a dominant population of immune cells in the GBM TME that contribute to most GBM hallmarks, including immunosuppression. The understanding of TAMs in GBM has been limited by the lack of powerful tools to characterize them. However, recent progress on single-cell technologies offers an opportunity to precisely characterize TAMs at the single-cell level and identify new TAM subpopulations with specific tumor-modulatory functions in GBM. In this Review, we discuss TAM heterogeneity and plasticity in the TME and summarize current TAM-targeted therapeutic potential in GBM. We anticipate that the use of single-cell technologies followed by functional studies will accelerate the development of novel and effective TAM-targeted therapeutics for GBM patients.
Figures