Evaluation of SARS-CoV-2-Specific T-Cell Activation with a Rapid On-Chip IGRA
- PMID: 36595218
- PMCID: PMC9878992
- DOI: 10.1021/acsnano.2c09018
Evaluation of SARS-CoV-2-Specific T-Cell Activation with a Rapid On-Chip IGRA
Abstract
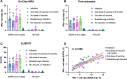
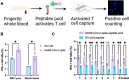
Interferon-gamma release assays (IGRAs) that measure pathogen-specific T-cell response rates can provide a more reliable estimate of protection than specific antibody levels but have limited potential for widespread use due to their workflow, personnel, and instrumentation demands. The major vaccines for SARS-CoV-2 have demonstrated substantial efficacy against all of its current variants, but approaches are needed to determine how these vaccines will perform against future variants, as they arise, to inform vaccine and public health policies. Here we describe a rapid, sensitive, nanolayer polylysine-integrated microfluidic chip IGRA read by a fluorescent microscope that has a 5 h sample-to-answer time and uses ∼25 μL of a fingerstick whole blood sample. Results from this assay correlated with those of a comparable clinical IGRA when used to evaluate the T-cell response to SARS-CoV-2 peptides in a population of vaccinated and/or infected individuals. Notably, this streamlined and inexpensive assay is suitable for high-throughput analyses in resource-limited settings for other infectious diseases.
Keywords: COVID-19; COVID-19 vaccine; IGRA; T-cell response; rapid test; whole blood assay.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Wilhelm A.; Widera M.; Grikscheit K.; Toptan T.; Schenk B.; Pallas C.; Metzler M.; Kohmer N.; Hoehl S.; Marschalek R. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. eBioMedicine 2022, 82, 104158. 10.1016/j.ebiom.2022.104158. - DOI - PMC - PubMed
-
- Singer S. R.; Angulo F. J.; Swerdlow D. L.; McLaughlin J. M.; Hazan I.; Ginish N.; Anis E.; Mendelson E.; Mor O.; Zuckerman N. S. Effectiveness of BNT162b2 mRNA COVID-19 vaccine against SARS-CoV-2 variant Beta (B.1.351) among persons identified through contact tracing in Israel: A prospective cohort study. eClinicalMedicine 2021, 42, 101190. 10.1016/j.eclinm.2021.101190. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

