Proteome alterations in erythrocytes with PIEZO1 gain-of-function mutations
- PMID: 36595486
- PMCID: PMC10333744
- DOI: 10.1182/bloodadvances.2022008673
Proteome alterations in erythrocytes with PIEZO1 gain-of-function mutations
Abstract
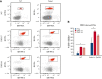
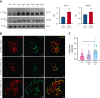
Gain-of-function mutations in PIEZO1 cause dehydrated hereditary stomatocytosis (DHS) or hereditary xerocytosis, an autosomal dominant hemolytic anemia characterized by high reticulocyte count, a tendency to macrocytosis, and mild jaundice, as well as by other variably penetrant clinical features, such as perinatal edema, severe thromboembolic complications after splenectomy, and hepatic iron overload. PIEZO1 mutations in DHS lead to slowed inactivation kinetics of the ion channel and/or facilitation of channel opening in response to physiological stimuli. To characterize the alterations of red blood cell proteome in patients with mutated PIEZO1, we used a differential approach to compare the proteome of patients with DHS (16 patients from 13 unrelated ancestries) vs healthy individuals. We identified new components in the regulation of the complex landscape of erythrocytes ion and volume balance mediated by PIEZO1. Specifically, the main impaired processes in patients with DHS were ion homeostasis, transmembrane transport, regulation of vesicle-mediated transport, and the proteasomal catabolic process. Functional assays demonstrated coexpression of PIEZO1 and band 3 when PIEZO1 was activated. Moreover, the alteration of the vesicle-mediated transport was functionally demonstrated by an increased vesiculation rate in patients with DHS compared with healthy controls. This finding also provides an explanation of the pathogenetic mechanism underlying the increased thrombotic rate observed in these patients. Finally, the newly identified proteins, involved in the intracellular signaling pathways altered by PIEZO1 mutations, could be used in the future as potential druggable targets in DHS.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures