Deep-sequencing of viral genomes from a large and diverse cohort of treatment-naive HIV-infected persons shows associations between intrahost genetic diversity and viral load
- PMID: 36595537
- PMCID: PMC9838853
- DOI: 10.1371/journal.pcbi.1010756
Deep-sequencing of viral genomes from a large and diverse cohort of treatment-naive HIV-infected persons shows associations between intrahost genetic diversity and viral load
Abstract
Background: Infection with human immunodeficiency virus type 1 (HIV) typically results from transmission of a small and genetically uniform viral population. Following transmission, the virus population becomes more diverse because of recombination and acquired mutations through genetic drift and selection. Viral intrahost genetic diversity remains a major obstacle to the cure of HIV; however, the association between intrahost diversity and disease progression markers has not been investigated in large and diverse cohorts for which the majority of the genome has been deep-sequenced. Viral load (VL) is a key progression marker and understanding of its relationship to viral intrahost genetic diversity could help design future strategies for HIV monitoring and treatment.
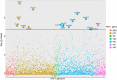
Methods: We analysed deep-sequenced viral genomes from 2,650 treatment-naive HIV-infected persons to measure the intrahost genetic diversity of 2,447 genomic codon positions as calculated by Shannon entropy. We tested for associations between VL and amino acid (AA) entropy accounting for sex, age, race, duration of infection, and HIV population structure.
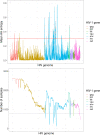
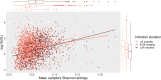
Results: We confirmed that the intrahost genetic diversity is highest in the env gene. Furthermore, we showed that mean Shannon entropy is significantly associated with VL, especially in infections of >24 months duration. We identified 16 significant associations between VL (p-value<2.0x10-5) and Shannon entropy at AA positions which in our association analysis explained 13% of the variance in VL. Finally, equivalent analysis based on variation in HIV consensus sequences explained only 2% of VL variance.
Conclusions: Our results elucidate that viral intrahost genetic diversity is associated with VL and could be used as a better disease progression marker than HIV consensus sequence variants, especially in infections of longer duration. We emphasize that viral intrahost diversity should be considered when studying viral genomes and infection outcomes.
Trial registration: Samples included in this study were derived from participants who consented in the clinical trial, START (NCT00867048) (23), run by the International Network for Strategic Initiatives in Global HIV Trials (INSIGHT). All the participant sites are listed here: http://www.insight-trials.org/start/my_phpscript/participating.php?by=site.
Copyright: © 2023 Gabrielaite et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures