Progressive transformation of the HIV-1 reservoir cell profile over two decades of antiviral therapy
- PMID: 36596305
- PMCID: PMC9839361
- DOI: 10.1016/j.chom.2022.12.002
Progressive transformation of the HIV-1 reservoir cell profile over two decades of antiviral therapy
Abstract
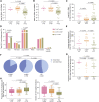
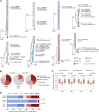
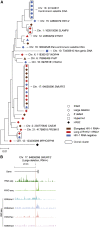
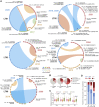
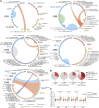
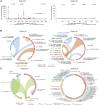
HIV-1 establishes a life-long reservoir of virally infected cells which cannot be eliminated by antiretroviral therapy (ART). Here, we demonstrate a markedly altered viral reservoir profile of long-term ART-treated individuals, characterized by large clones of intact proviruses preferentially integrated in heterochromatin locations, most prominently in centromeric satellite/micro-satellite DNA. Longitudinal evaluations suggested that this specific reservoir configuration results from selection processes that promote the persistence of intact proviruses in repressive chromatin positions, while proviruses in permissive chromosomal locations are more likely to be eliminated. A bias toward chromosomal integration sites in heterochromatin locations was also observed for intact proviruses in study participants who maintained viral control after discontinuation of antiretroviral therapy. Together, these results raise the possibility that antiviral selection mechanisms during long-term ART may induce an HIV-1 reservoir structure with features of deep latency and, possibly, more limited abilities to drive rebound viremia upon treatment interruptions.
Keywords: FLIP-seq; HIV; HIV cure; HIV eradication; MIP-seq; block and lock; integration sites; latency; post-treatment controllers; retroviral pathogenesis.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R., et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. - PubMed
-
- Wong J.K., Hezareh M., Günthard H.F., Havlir D.V., Ignacio C.C., Spina C.A., Richman D.D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. - PubMed
-
- Yukl S.A., Kaiser P., Kim P., Telwatte S., Joshi S.K., Vu M., Lampiris H., Wong J.K. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018;10:eaap9927. doi: 10.1126/scitranslmed.aap9927. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- UM1 AI069494/AI/NIAID NIH HHS/United States
- K24 AI155233/AI/NIAID NIH HHS/United States
- R01 HL134539/HL/NHLBI NIH HHS/United States
- UM1 AI164570/AI/NIAID NIH HHS/United States
- R01 AI130005/AI/NIAID NIH HHS/United States
- R01 DK120387/DK/NIDDK NIH HHS/United States
- UM1 AI164560/AI/NIAID NIH HHS/United States
- UM1 AI164566/AI/NIAID NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
- R33 DA047034/DA/NIDA NIH HHS/United States
- R01 AI152979/AI/NIAID NIH HHS/United States
- UM1 AI069412/AI/NIAID NIH HHS/United States
- UM1 AI068634/AI/NIAID NIH HHS/United States
- R61 DA047034/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

