The ensemble of gene regulatory networks at mutation-selection balance
- PMID: 36596452
- PMCID: PMC9810427
- DOI: 10.1098/rsif.2022.0075
The ensemble of gene regulatory networks at mutation-selection balance
Abstract
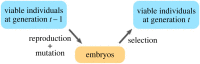
The evolution of diverse phenotypes both involves and is constrained by molecular interaction networks. When these networks influence patterns of expression, we refer to them as gene regulatory networks (GRNs). Here, we develop a model of GRN evolution analogous to work from quasi-species theory, which is itself essentially the mutation-selection balance model from classical population genetics extended to multiple loci. With this GRN model, we prove that-across a broad spectrum of selection pressures-the dynamics converge to a stationary distribution over GRNs. Next, we show from first principles how the frequency of GRNs at equilibrium is related to the topology of the genotype network, in particular, via a specific network centrality measure termed the eigenvector centrality. Finally, we determine the structural characteristics of GRNs that are favoured in response to a range of selective environments and mutational constraints. Our work connects GRN evolution to quasi-species theory-and thus to classical populations genetics-providing a mechanistic explanation for the observed distribution of GRNs evolving in response to various evolutionary forces, and shows how complex fitness landscapes can emerge from simple evolutionary rules.
Keywords: gene regulatory networks; mutation–selection balance; neutral network; quasi-species theory.
Figures



References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous
