Protein-protein complexes can undermine ultrasensitivity-dependent biological adaptation
- PMID: 36596458
- PMCID: PMC9810431
- DOI: 10.1098/rsif.2022.0553
Protein-protein complexes can undermine ultrasensitivity-dependent biological adaptation
Abstract
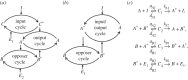
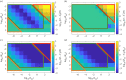
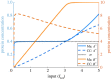
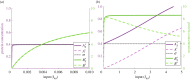
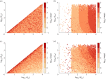
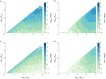
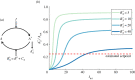
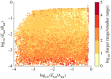
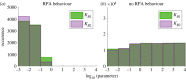
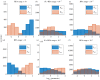
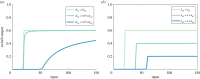
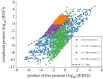
Robust perfect adaptation (RPA) is a ubiquitously observed signalling response across all scales of biological organization. A major class of network architectures that drive RPA in complex networks is the Opposer module-a feedback-regulated network into which specialized integral-computing 'opposer node(s)' are embedded. Although ultrasensitivity-generating chemical reactions have long been considered a possible mechanism for such adaptation-conferring opposer nodes, this hypothesis has relied on simplified Michaelian models, which neglect the presence of protein-protein complexes. Here we develop complex-complete models of interlinked covalent-modification cycles with embedded ultrasensitivity, explicitly capturing all molecular interactions and protein complexes. Strikingly, we demonstrate that the presence of protein-protein complexes thwarts the network's capacity for RPA in any 'free' active protein form, conferring RPA capacity instead on the concentration of a larger protein pool consisting of two distinct forms of a single protein. We further show that the presence of enzyme-substrate complexes, even at comparatively low concentrations, play a crucial and previously unrecognized role in controlling the RPA response-significantly reducing the range of network inputs for which RPA can obtain, and imposing greater parametric requirements on the RPA response. These surprising results raise fundamental new questions as to the biochemical requirements for adaptation-conferring Opposer modules within complex cellular networks.
Keywords: Michaelis–Menten; chemical reaction networks; feedback; mass-action; robust perfect adaptation; ultrasensitivity.
Figures


References
-
- Araujo RP, Vittadello ST, Stumpf MPH. 2021. Bayesian and algebraic strategies to design in synthetic biology. Proc. IEEE 110, 675-687. ( 10.1109/JPROC.2021.3129527) - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
