Genome wide association analysis of root hair traits in rice reveals novel genomic regions controlling epidermal cell differentiation
- PMID: 36597029
- PMCID: PMC9811729
- DOI: 10.1186/s12870-022-04026-5
Genome wide association analysis of root hair traits in rice reveals novel genomic regions controlling epidermal cell differentiation
Abstract
Background: Genome wide association (GWA) studies demonstrate linkages between genetic variants and traits of interest. Here, we tested associations between single nucleotide polymorphisms (SNPs) in rice (Oryza sativa) and two root hair traits, root hair length (RHL) and root hair density (RHD). Root hairs are outgrowths of single cells on the root epidermis that aid in nutrient and water acquisition and have also served as a model system to study cell differentiation and tip growth. Using lines from the Rice Diversity Panel-1, we explored the diversity of root hair length and density across four subpopulations of rice (aus, indica, temperate japonica, and tropical japonica). GWA analysis was completed using the high-density rice array (HDRA) and the rice reference panel (RICE-RP) SNP sets.
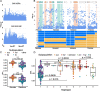
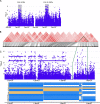
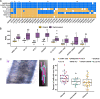
Results: We identified 18 genomic regions related to root hair traits, 14 of which related to RHD and four to RHL. No genomic regions were significantly associated with both traits. Two regions overlapped with previously identified quantitative trait loci (QTL) associated with root hair density in rice. We identified candidate genes in these regions and present those with previously published expression data relevant to root hair development. We re-phenotyped a subset of lines with extreme RHD phenotypes and found that the variation in RHD was due to differences in cell differentiation, not cell size, indicating genes in an associated genomic region may influence root hair cell fate. The candidate genes that we identified showed little overlap with previously characterized genes in rice and Arabidopsis.
Conclusions: Root hair length and density are quantitative traits with complex and independent genetic control in rice. The genomic regions described here could be used as the basis for QTL development and further analysis of the genetic control of root hair length and density. We present a list of candidate genes involved in root hair formation and growth in rice, many of which have not been previously identified as having a relation to root hair growth. Since little is known about root hair growth in grasses, these provide a guide for further research and crop improvement.
Keywords: Candidate gene; GWAS; Rice; Root biology; Root hairs; Subpopulation.
© 2023. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures










Similar articles
-
Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping.PLoS Genet. 2011 Aug;7(8):e1002221. doi: 10.1371/journal.pgen.1002221. Epub 2011 Aug 4. PLoS Genet. 2011. PMID: 21829395 Free PMC article.
-
Validation of Yield Component Traits Identified by Genome-Wide Association Mapping in a tropical japonica × tropical japonica Rice Biparental Mapping Population.Plant Genome. 2019 Mar;12(1). doi: 10.3835/plantgenome2018.04.0021. Plant Genome. 2019. PMID: 30951093
-
Genome-Wide Association Study of Root System Development at Seedling Stage in Rice.Genes (Basel). 2020 Nov 25;11(12):1395. doi: 10.3390/genes11121395. Genes (Basel). 2020. PMID: 33255557 Free PMC article.
-
Genomics of Treatable Traits in Asthma.Genes (Basel). 2023 Sep 20;14(9):1824. doi: 10.3390/genes14091824. Genes (Basel). 2023. PMID: 37761964 Free PMC article. Review.
-
Use of cell lines in the investigation of pharmacogenetic loci.Curr Pharm Des. 2009;15(32):3782-95. doi: 10.2174/138161209789649475. Curr Pharm Des. 2009. PMID: 19925429 Free PMC article. Review.
Cited by
-
DIRT/µ: automated extraction of root hair traits using combinatorial optimization.J Exp Bot. 2025 Jan 10;76(2):285-298. doi: 10.1093/jxb/erae385. J Exp Bot. 2025. PMID: 39269014 Free PMC article.
-
Genome- and Transcriptome-wide Association Studies to Discover Candidate Genes for Diverse Root Phenotypes in Cultivated Rice.Rice (N Y). 2023 Dec 8;16(1):55. doi: 10.1186/s12284-023-00672-x. Rice (N Y). 2023. PMID: 38063928 Free PMC article.
References
-
- Dittmer HJ. A quantitative study of the roots and root hairs of a winter rye plant (Secale cereale) Am J Bot. 1937;24:417–420. doi: 10.1002/j.1537-2197.1937.tb09121.x. - DOI
-
- Misra RK, Alston AM, Dexter AR. Role of root hairs in phosphorus depletion from a macrostructured soil. Plant Soil. 1988;107:11–18. doi: 10.1007/BF02371538. - DOI
-
- Segal E, Kushnir T, Mualem Y, Shani U. Water uptake and hydraulics of the root hair rhizosphere. Vadose Zone J. 2008;7:1027. doi: 10.2136/vzj2007.0122. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

