Trends in carbapenem resistance in Pre-COVID and COVID times in a tertiary care hospital in North India
- PMID: 36597098
- PMCID: PMC9808733
- DOI: 10.1186/s12941-022-00549-9
Trends in carbapenem resistance in Pre-COVID and COVID times in a tertiary care hospital in North India
Abstract
Background: Carbapenem resistance is endemic in the Indian sub-continent. In this study, carbapenem resistance rates and the prevalence of different carbapenemases were determined in Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa during two periods; Pre-COVID (August to October 2019) and COVID (January to February 2021) in a north-Indian tertiary care hospital.
Methods: Details of patient demographics and clinical condition was collated from the Hospital Information System and detection of carbapenemases NDM, OXA-48, VIM, IMP and KPC was done by Polymerase chain reaction (PCR) in 152 and 138 non-consecutive carbapenem resistant isolates during the two study periods respectively. Conjugation assay and sequencing of NDM and OXA-48 gene was done on a few selected isolates.
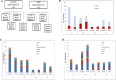
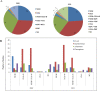
Results: As compared to Pre-COVID period, co-morbidities and the mortality rates were higher in patients harbouring carbapenem resistant organisms during the COVID period. The overall carbapenem resistance rate for all the four organisms increased from 23 to 41% between the two periods of study; with Pseudomonas aeruginosa and Klebsiella pneumoniae showing significant increase (p < 0.05). OXA-48, NDM and co-expression of NDM and OXA-48 were the most common genotypes detected. NDM-5 and OXA-232 were most common variants of NDM and OXA-48 family respectively during both the study periods.
Conclusion: Higher rate of carbapenem resistance in COVID times could be attributed to increase in number of patients with co-morbidities. However, genetic elements of carbapenem resistance largely remained the same in the two time periods.
Keywords: COVID; Carbapenem resistance; India; NDM; OXA-48 family.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Chandy SJ, Thomas K, Mathai E, Antonisamy B, Holloway KA, Stalsby LC. Patterns of antibiotic use in the community and challenges of antibiotic surveillance in a lower-middle-income country setting: a repeated cross-sectional study in Vellore, South India. J Antimicrob Chemother. 2013;68(1):229–236. doi: 10.1093/jac/dks355. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

